HW2. Signal Processing
Topics: Image blending, Fourier transform, Image compression
import numpy as np, matplotlib as mpl, matplotlib.pyplot as plt, os
import urllib.request
import math
from PIL import Image
import scipy.ndimage # For image filtering
from scipy import signal
from scipy import fft
import imageio # For loading images
import cv2
Problem 1: Image blending
1.(a) Reconstructing an image
In this problem, we construct a Laplacian pyramid with 4 levels. We use the Laplacian pyramid to reconstruct the original image.
img_url = 'https://www.eecs.umich.edu/courses/eecs442-ahowens/pset_images/ps2/bbb-1000.jpg'
with open('bbb.jpg', 'wb') as out:
out.write(urllib.request.urlopen(img_url).read())
img = Image.open('bbb.jpg')
img = img.resize((400,200))
img=np.array(img)
plt.figure()
plt.title('Input image')
plt.axis('off')
plt.imshow(img)
<matplotlib.image.AxesImage at 0x7fd4b21a8c50>

First, we will implement the following functions, which will be used to create a Laplacian pyramid from an image, and to reconstruct an image from a Laplacian pyramid:
- pyramid upsample
- pyramid downsample
- gen gaussian pyramid
- gen laplacian pyramid
- reconstruct img
In your implementation, use Gaussian kernels for pyramid upsample and pyramid downsample. The kernel for pyramid upsample should be the same as the one for pyramid downsample, except that it will be multiplied by 4. Use a Laplacian pyramid with 4 levels, and set the standard deviation of the Gaussian kernel as $\sigma = 1$. Please plot the original image, the Lapla- cian pyramid, and the reconstructed image. (Hint: np.insert may come in handy when implementing pyramid upsample.)
def calculate_gaussian_value(x, y, sigma):
x1 = 2 * np.pi * (sigma**2)
x2 = np.exp(-(x**2 + y**2) / (2 * sigma **2))
return (1 / x1) * x2
def make_gaussian_filter(sigma, kernel_size):
gaussian_filter = np.zeros(kernel_size)
for i in range(gaussian_filter.shape[0]):
for j in range(gaussian_filter.shape[1]):
gaussian_filter[i, j] = calculate_gaussian_value(i - kernel_size[0]//2, j - kernel_size[1]//2, sigma)
gaussian_filter /= np.sum(gaussian_filter)
return gaussian_filter
def pyramid_upsample(img, kernel_size=(5,5)):
"""
Upsamples the given pyramid image.
Input:
- img: an image of shape M x N x C
- kernel_size: a tuple representing the shape of the 2D kernel
Returns:
- upsampled: an image represented as an array of shape 2M x 2N x C
"""
#############################################################################
# TODO: Implement pyramid upsampling. #
#############################################################################
x_upsampled = [np.insert(img[:, :, c], np.arange(0, img.shape[0]), 0, axis = 0) for c in range(img.shape[2])]
xy_upsampled = [np.insert(x_upsampled[c], np.arange(0, img.shape[1]), 0, axis = 1) for c in range(img.shape[2])]
gaussian_filter = make_gaussian_filter(1, kernel_size)
gaussian_filter *= 4
channel_blurred = [scipy.ndimage.convolve(xy_upsampled[c], gaussian_filter) for c in range(img.shape[2])]
upsampled = np.dstack(channel_blurred)
#############################################################################
# END OF YOUR CODE #
#############################################################################
return upsampled
def pyramid_downsample(img, kernel_size=(5,5)):
"""
Downsamples the given pyramid image.
Input:
- img: an image of shape M x N x C
- kernel_size: a tuple representing the shape of the 2D kernel
Returns:
- downsampled: an image of shape M/2 x N/2 x C
"""
#############################################################################
# TODO: Implement pyramid downsampling. #
#############################################################################
gaussian_filter = make_gaussian_filter(1, kernel_size)
channel_blurred = [scipy.ndimage.convolve(img[:, :, c], gaussian_filter) for c in range(img.shape[2])]
channel_blurred_x_downsampled = [np.delete(channel_blurred[c], np.arange(1, img.shape[0], 2), axis = 0) for c in range(len(channel_blurred))]
channel_blurred_xy_downsampled = [np.delete(channel_blurred_x_downsampled[c], np.arange(1, img.shape[1], 2), axis = 1) for c in range(len(channel_blurred_x_downsampled))]
downsampled = np.dstack(channel_blurred_xy_downsampled)
#############################################################################
# END OF YOUR CODE #
#############################################################################
return downsampled
def gen_gaussian_pyramid(img, num_levels):
"""
Generates an entire Gaussian pyramid.
Input:
- img: an image of shape M x N x C
- num_levels: number of levels in the Gaussian pyramid
Return:
- gp: list, the generated levels (imgs) of the Gaussian pyramid
"""
#############################################################################
# TODO: Construct a Gaussian pyramid given a base image `img`. #
#############################################################################
gp = [img]
for level in range(num_levels - 1):
gp.append(pyramid_downsample(gp[level]))
#############################################################################
# END OF YOUR CODE #
#############################################################################
return gp
def gen_laplacian_pyramid(gp, num_levels):
"""
Generates an entire Laplacian pyramid.
Input:
gp: list, levels of a Gaussian pyramid
Return:
lp: list, the generated levels (imgs) of the Laplacian pyramid
"""
#############################################################################
# TODO: Construct a Laplacian pyramid given final level of the Gaussian pyramid. #
# The 0th element of lp should be the output of the Gaussian pyramid. #
#############################################################################
lp = [gp[num_levels - 1]]
for level in np.arange(len(gp) - 1, 0, -1):
lp_image = pyramid_upsample(gp[level])
lp.append(gp[level - 1] - lp_image)
#############################################################################
# END OF YOUR CODE #
#############################################################################
return lp
def reconstruct_img(lp):
"""
Reconstructs an image using a laplacian pyramid.
Input:
lp: list, levels (imgs) of a Laplacian pyramid
Return:
recon_img: reconstructed image
"""
recon_img = lp[0]
for i in range(1, len(lp)):
###########################################################################
# TODO: For each level, reconstruct the image from the Laplacian pyramid. #
###########################################################################
recon_img = lp[i] + pyramid_upsample(recon_img)
###########################################################################
# END OF YOUR CODE #
###########################################################################
return recon_img
# Run above functions and visualize results on bbb.jpg
img = img.astype('float')
num_levels = 4 # including the original img
gp = gen_gaussian_pyramid(img, num_levels)
lp = gen_laplacian_pyramid(gp, num_levels)
recon_img = reconstruct_img(lp)
print("Gaussian pyramid:")
for i, level in enumerate(gp):
level=(level-np.amin(level))/(np.amax(level)-np.amin(level))
plt.figure(figsize=[plt.rcParams.get('figure.figsize')[0]/(2**i),plt.rcParams.get('figure.figsize')[1]/(2**i)])
plt.axis('off')
print("Level {}:".format(i),level.shape)
plt.imshow(level)
plt.show()
print("Laplacian pyramid:")
for i, level in enumerate(lp):
level_plot=level.clip(0, 255).astype('uint8')
plt.figure(figsize=[plt.rcParams.get('figure.figsize')[0]/(2**(num_levels-i-1)),plt.rcParams.get('figure.figsize')[1]/(2**(num_levels-i-1))])
plt.axis('off')
print("Level {}:".format(i),level.shape)
plt.imshow(level_plot)
plt.show()
print("")
img=img.astype(np.uint8)
plt.figure()
plt.title("Original Image")
plt.axis('off')
plt.imshow(img)
plt.show()
recon_img=recon_img.astype(np.uint8)
plt.figure()
plt.title("Reconstructed Image")
plt.axis('off')
plt.imshow(recon_img)
plt.show()
Gaussian pyramid:
Level 0: (200, 400, 3)

Level 1: (100, 200, 3)

Level 2: (50, 100, 3)

Level 3: (25, 50, 3)

Laplacian pyramid:
Level 0: (25, 50, 3)

Level 1: (50, 100, 3)

Level 2: (100, 200, 3)

Level 3: (200, 400, 3)


1.(b) Blending two images
In this problem, we blend an image of an orange and an apple.
Implement the function pyramid_blend(im1, im2, mask, num levels). Its inputs are two images and a binary mask (indicating which pixels to use from each image). The function produces a Laplacian pyramid with num_levels levels that will blend the two image inputs. Use your function to blend the images of an orange and an apple that we provided in the Colab notebook. Plot the blended images with num_levels $\in$ {1, 2, 3, 4, 5, 6}. Please describe the difference between the blended images as the number of levels in the Laplacian pyramid varies: how does the result change as we increase the number of levels?
To obtain color images, you can apply the blending to each color channel independently. In our implementation, this did not require any extra code (the same code worked on single-channel and multi-channel images due to numpy broadcasting). However, your implementation may differ.
# Download the `orange.jpg` and `apple.jpg` images.
base_url = 'https://inst.eecs.berkeley.edu/~cs194-26/fa17/upload/files/proj3/cs194-26-acr/img/multires1/'
for name in ['orange.jpg', 'apple.jpg']:
with open(name, 'wb') as out:
url = os.path.join(base_url, name)
out.write(urllib.request.urlopen(url).read())
# Resize images and create the image mask.
img1 = Image.open('apple.jpg')
img2 =Image.open('orange.jpg')
width,height=img1.size
new_width,new_height=width//2,height//2
img1 = img1.resize((new_width,new_height))
img1=np.array(img1)
img2 = img2.resize((new_width,new_height))
img2=np.array(img2)
mask = np.zeros_like(img1)
mask[:,:mask.shape[1]//2, :] = 1
def pyramid_blend(img1, img2, mask, num_levels=6):
"""
This function produces the Laplacian pyramid blend of two images.
Input:
- img1: N x M x C uint8 array image
- img2: N x M x C uint8 array image
- mask: N x M array, all elements are either 0s or 1s
- num_levels: int, height of the pyramid
Return:
- img_blend: the blended image, an N x M x C uint8 array
"""
# Build Gaussian pyramids for img1, img2, and mask
gp1, gp2, gpm = gen_gaussian_pyramid(img1, num_levels), gen_gaussian_pyramid(img2, num_levels), gen_gaussian_pyramid(mask, num_levels)
# Build Laplacian pyramids for img1 and img2
lp1, lp2, lpm = gen_laplacian_pyramid(gp1, num_levels), gen_laplacian_pyramid(gp2, num_levels), gen_laplacian_pyramid(gpm, num_levels)
#############################################################################
# TODO: Construct the Laplacian pyramid and use it to blend the images. #
#############################################################################
lp_blended = []
gpm.reverse()
for i in range(num_levels):
lp_blended.append(np.multiply(lp1[i], gpm[i]) + np.multiply(lp2[i], 1 - gpm[i]))
img_blend = reconstruct_img(lp_blended)
#############################################################################
# END OF YOUR CODE #
#############################################################################
return img_blend
# Display images and mask
# Plotting image 1 - Apple
plt.figure()
plt.title('Image 1')
plt.axis('off')
plt.imshow(img1)
# Plotting image 2 - Orange
plt.figure()
plt.title('Image 2')
plt.axis('off')
plt.imshow(img2)
# Plotting the mask
plt.figure()
plt.title('Mask')
plt.axis('off')
plt.imshow(mask * 255)
<matplotlib.image.AxesImage at 0x7fd4b184fd50>



# Visualize Laplacian pyramid blend by varying number of levels
for n_l in range(1, 7):
img_blend = pyramid_blend(img1.astype(float), img2.astype(float), mask.astype(float), num_levels=n_l)
img_blend=img_blend.astype(np.uint8)
plt.figure()
plt.title('Number of levels = {}'.format(n_l))
plt.axis('off')
plt.imshow(img_blend)






Problem 2: 2D DFT and the Convolution Theorem
2.(a) Frequence domain
In this problem, we compare the output of a Gaussian filter through i) direct convolution in the spatial domain and ii) multiplication in the frequency domain.
img_url = 'https://mapproxy.studentlife.umich.edu/images/electrical-engineering-and-computer-science-bld/XP2S3935.jpg'
with open('eecs.jpg', 'wb') as out:
out.write(urllib.request.urlopen(img_url).read())
img=Image.open('eecs.jpg').convert('L') #Here, we are converting the RGB image into a grayscale
width,height=img.size
new_width,new_height=width//2,height//2
img = img.resize((new_width,new_height))
img=np.array(img)
plt.figure()
plt.imshow(img)
plt.title('Original image')
plt.show()
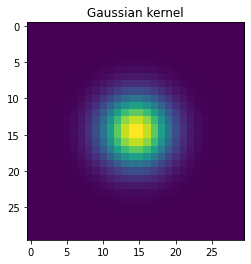
# Make a 2-D kernel Gaussian kernel
t = np.linspace(-10, 10, 30)
bump = np.exp(-0.1*t**2)
bump /= np.trapz(bump) # normalize the integral to 1
kernel = bump[:, np.newaxis] * bump[np.newaxis, :]
plt.imshow(kernel)
plt.title('Gaussian kernel')
plt.show()
# Convolution in spatial domain
#############################################################################
# TODO: Convolve the gaussian filter with the image in the spatial domain. #
#############################################################################
img1 = scipy.signal.convolve2d(img, kernel)
#############################################################################
# END OF YOUR CODE #
#############################################################################
plt.figure()
plt.title('Direct convolution')
plt.imshow(img1, cmap='gray')
plt.show()
# Multiplication in frequency domain
# This shape needs to be passed into the fft2 function
shape = (img.shape[0]+kernel.shape[0]-1, img.shape[1]+kernel.shape[1]-1)
#############################################################################
# TODO: Use the convolution theorem to convolve the provided image with #
# a Gaussian filter. #
#############################################################################
img_transformed = scipy.fft.fft2(img, shape)
kernel_transformed = scipy.fft.fft2(kernel, shape)
img2 = np.abs(scipy.fft.ifft2(np.multiply(img_transformed, kernel_transformed)))
#############################################################################
# END OF YOUR CODE #
#############################################################################
plt.figure()
plt.title('Multiplication in frequency domain')
plt.imshow(img2, cmap='gray')
plt.show()



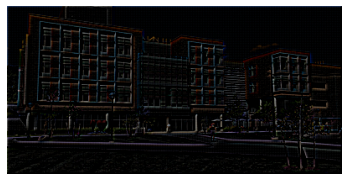
Problem 3 JPEG: Image Compression
The Central Campus dean has escalated his mysterious feud with the JPEG Standards Committee: beginning next week, UMich will, by his order, be entirely JPEG-free. Fearing the huge increase in storage costs from dealing with uncompressed images, U-M ITS has created a simplified version of JPEG that they plan to deploy to all of their data centers over the weekend. They have turned to you for help in completing one small component of the system: the frequency decomposition.
JPEG image compression takes advantage of the fact that human vision is less sensitive to high frequency components than to low frequency components. It is therefore possible to discard some high frequency components without significantly reducing the visual image quality. In this problem, you will implement a variation of the Fourier transform called the discrete cosine transform (DCT), and explore it via simple visualizations. After implementing the DCT, you can plug it into U-M ITS’s new compression system to confirm that the model leads to significantly smaller images.
Note: Despite the large amount of code we have given you for the JPEG compression system (and the length of the question), this problem should only require a few lines of code!.
JPEG image compression can be divided into several steps:
- Break the image into tiny 8 × 8 patches.
- For each patch, apply the Discrete cosine transform (DCT) to obtain a frequency decomposition.
- Quantization: high frequency DCT coefficients with small magnitude are set to zero.
- Finally, we losslessly compress the quantized DCT coefficients. Since these contain a large number of zeros, they will compress much more easily than before quantization. This step uses Huffman coding (see here for more information, if you are interested)
Recovering the image from the encoded file follows the reverse of the above steps:
- Decoding the Huffman-coded file to get the quantized DCT coefficients
- De-quantization, where quantized DCT coefficients are scaled back to the actual scale
- Inverse DCT to reconstruct the image.
By following the above steps, we will compress a grayscale image into a compact encoded file, examine its new file size, and then decode that compact file and reconstruct the image. You will implement the DCT transform part (step 1). We have implemented the remaining code for you. Please note that the amount of code that you’ll need to write is quite minimal!
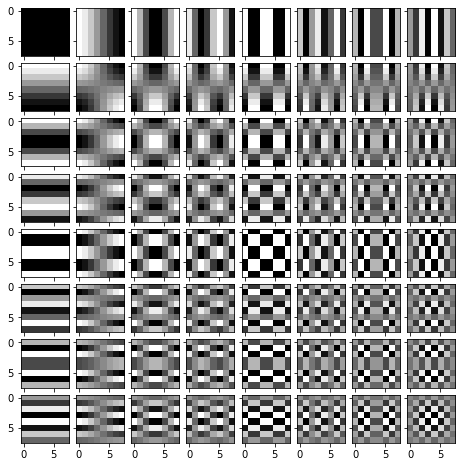
3.(a) Basis for 2D Discrete Cosine Transform
A core component of 2D DCT is the 2D DCT basis, which is a set of 2D sinusoidal images with different spatial frequencies of the form $cos\frac{\pi (2m + 1) p}{2M}cos\frac{\pi (2n + 1)q}{2N}$. Implement function build 2D DCT basis that will compute this basis image set. Visualize the basis image set using the given code.
import numpy as np
def build_2D_DCT_basis(Nx, Ny):
#Create 2D coordinate grid
X = np.arange(Nx)[None,:].repeat(Ny,axis=0)
Y = np.arange(Ny)[:,None].repeat(Nx,axis=1)
DCT_basis = np.zeros([Ny,Nx,Ny,Nx]) # First two indices correspond to frequency components (ky,kx), last two indices correspond to spatial location (y,x).
for kx in range(Nx):
for ky in range(Ny):
#############################################################################
# TODO: Fill in DCT_basis's frequency components as defined in the HW. #
#############################################################################
for x in range(DCT_basis[kx][ky].shape[1]):
for y in range(DCT_basis[kx][ky].shape[0]):
DCT_basis[ky, kx, y, x] = np.cos((np.pi * (2* x + 1) * kx) / (2 * DCT_basis[kx][ky].shape[1])) * np.cos((np.pi * (2* y + 1) * ky) / (2 * DCT_basis[kx][ky].shape[0]))
#############################################################################
# END OF YOUR CODE #
#############################################################################
return DCT_basis
# Build the DCT basis and visualize it.
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import ImageGrid
Nx, Ny = 8, 8
DCT_basis = build_2D_DCT_basis(Nx, Ny)
fig = plt.figure(figsize=(8., 8.))
grid = ImageGrid(fig, 111, # similar to subplot(111)
nrows_ncols=(Ny, Nx), # creates a grid of axes
axes_pad=0.1, # pad between axes in inch.
)
for ax, im in zip(grid, DCT_basis.reshape(-1,Ny,Nx)):
# Iterating over the grid returns the Axes.
ax.imshow(im, cmap = 'gray')
plt.show()
3.(b) 2D Discrete Cosine Transform
DCT 2D and IDCT 2D for performing 2D DCT and inverse 2D DCT are implemented using the built 2D DCT basis, read through the code of these two functions and make sure you understand them.
def DCT_2D(image_patch):
"""
2D DCT transform of image patch of shape (H,W)
"""
H,W = image_patch.shape
DCT_basis = build_2D_DCT_basis(W,H)
DCT_coeffcients = np.zeros([H,W])
for h in range(H):
for w in range(W):
norm_const_h = 1/np.sqrt(H) if h == 0 else np.sqrt(2/H)
norm_const_w = 1/np.sqrt(W) if w == 0 else np.sqrt(2/W)
DCT_coeffcients[h,w] = norm_const_h * norm_const_w * (DCT_basis[h, w] * image_patch).sum()
return DCT_coeffcients
def IDCT_2D(image_spectrum):
"""
2D inverse DCT trasform.
"""
H,W = image_spectrum.shape
DCT_basis = build_2D_DCT_basis(W,H)
image = np.zeros([H,W])
norm_matrix = 1/np.sqrt(H*W) * np.ones([H,W])
norm_matrix[1:,0] *= np.sqrt(2)
norm_matrix[0,1:] *= np.sqrt(2)
norm_matrix[1:,1:] *= 2
for h in range(H):
for w in range(W):
norm_const_h = 1/np.sqrt(H) if h == 0 else np.sqrt(2/H)
norm_const_w = 1/np.sqrt(W) if w == 0 else np.sqrt(2/W)
image += norm_const_h * norm_const_w * (DCT_basis[h, w] * image_spectrum[h,w])
return image
image_patch = np.random.rand(8,8)
# Verify above gives same result as using scipy fftpack
from scipy import fftpack
def dct_2d(image):
return fftpack.dct(fftpack.dct(image.T, norm='ortho').T, norm='ortho')
def idct_2d(image):
return fftpack.idct(fftpack.idct(image.T, norm='ortho').T, norm='ortho')
print(np.abs(DCT_2D(image_patch) - dct_2d(image_patch)).sum())
print(np.abs(IDCT_2D(image_patch) - idct_2d(image_patch)).sum())
4.006430603942235e-14
2.552472122552274e-14
3.(c) JPEG Image Compression
Now, the full JPEG compression system should be ready to run! In the encoding part of the code, try image compression using different quantization tables, including dc_only, first_3, first_6 and first_10. Each of these tables discards a different set of frequency components; tables that discard more components produce a more compressed, but lossier, image. For each quantization table, report the compress ratio (size of the compressed file divided by size of uncompressed file). Also, please briefly comment on their reconstructed image quality and file size, and explain why it is the case.
#Download the image to be compressed
!wget -O original.png "https://docs.google.com/uc?export=download&id=1Ww2zAALWCuOmDbYw1XM5ACmFgIA8u6_9"
--2022-09-22 02:52:04-- https://docs.google.com/uc?export=download&id=1Ww2zAALWCuOmDbYw1XM5ACmFgIA8u6_9
Resolving docs.google.com (docs.google.com)... 172.253.115.113, 172.253.115.101, 172.253.115.100, ...
Connecting to docs.google.com (docs.google.com)|172.253.115.113|:443... connected.
HTTP request sent, awaiting response... 303 See Other
Location: https://doc-0s-7s-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/p5fpmtt5cvjtvq5udvkln01ok2j83out/1663815075000/11328405491383495500/*/1Ww2zAALWCuOmDbYw1XM5ACmFgIA8u6_9?e=download&uuid=d80d3062-6ea6-488f-b7ff-637a369e8bf1 [following]
Warning: wildcards not supported in HTTP.
--2022-09-22 02:52:04-- https://doc-0s-7s-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/p5fpmtt5cvjtvq5udvkln01ok2j83out/1663815075000/11328405491383495500/*/1Ww2zAALWCuOmDbYw1XM5ACmFgIA8u6_9?e=download&uuid=d80d3062-6ea6-488f-b7ff-637a369e8bf1
Resolving doc-0s-7s-docs.googleusercontent.com (doc-0s-7s-docs.googleusercontent.com)... 172.253.122.132, 2607:f8b0:4004:c09::84
Connecting to doc-0s-7s-docs.googleusercontent.com (doc-0s-7s-docs.googleusercontent.com)|172.253.122.132|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 231014 (226K) [image/png]
Saving to: ‘original.png’
original.png 100%[===================>] 225.60K --.-KB/s in 0.002s
2022-09-22 02:52:05 (119 MB/s) - ‘original.png’ saved [231014/231014]
Utility functions
We will first define some utility functions. You don’t need to implement anything in this block or read all of its details.
The only important functions in this part you may want to look at to understand better are quantize and load_quantization_table.
# We will first define some utility functions. You don't need to implement anything in this block or read all of its details.
# The only important functions in this part you may want to look at to understand better are 'quantize' and 'load_quantization_table'.
# Code in this part is based on https://github.com/ghallak/jpeg-python
from queue import PriorityQueue
class HuffmanTree:
"""
Class for creating Huffman Table, given an input array. The huffman encodes
input data based on their appearance frequency,
i.e., assign a shorter code for more frequently appearing values.
"""
def __init__(self, arr):
q = PriorityQueue()
# calculate frequencies and insert them into a priority queue
for val, freq in self.__calc_freq(arr).items():
q.put(self.__Node.init_leaf(val, freq))
while q.qsize() >= 2:
u = q.get()
v = q.get()
q.put(self.__Node.init_node(u, v))
self.__root = q.get()
# dictionaries to store huffman table
self.__value_to_bitstring = dict()
def value_to_bitstring_table(self):
if len(self.__value_to_bitstring.keys()) == 0:
self.__create_huffman_table()
return self.__value_to_bitstring
def __create_huffman_table(self):
def tree_traverse(current_node, bitstring=''):
if current_node is None:
return
if current_node.is_leaf():
self.__value_to_bitstring[current_node.value] = bitstring
return
tree_traverse(current_node.left_child, bitstring + '0')
tree_traverse(current_node.right_child, bitstring + '1')
tree_traverse(self.__root)
def __calc_freq(self, arr):
freq_dict = dict()
for elem in arr:
if elem in freq_dict:
freq_dict[elem] += 1
else:
freq_dict[elem] = 1
return freq_dict
class __Node:
def __init__(self, value, freq, left_child, right_child):
self.value = value
self.freq = freq
self.left_child = left_child
self.right_child = right_child
@classmethod
def init_leaf(cls, value, freq):
return cls(value, freq, None, None)
@classmethod
def init_node(cls, left_child, right_child):
freq = left_child.freq + right_child.freq
return cls(None, freq, left_child, right_child)
def is_leaf(self):
return self.value is not None
def __eq__(self, other):
stup = self.value, self.freq, self.left_child, self.right_child
otup = other.value, other.freq, other.left_child, other.right_child
return stup == otup
def __nq__(self, other):
return not (self == other)
def __lt__(self, other):
return self.freq < other.freq
def __le__(self, other):
return self.freq < other.freq or self.freq == other.freq
def __gt__(self, other):
return not (self <= other)
def __ge__(self, other):
return not (self < other)
def load_quantization_table(component):
# Quantization tables determine how much DCT coefficents are divided during
# quantization. Larger value means it is more likely to be a 0 after division.
# Setting large values like 100000 means that frequency component is set to 0 for sure.
if component == 'dc_only':
q = np.array([[1, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000]])
elif component == 'first_3':
q = np.array([[1, 1, 100000, 100000, 100000, 100000, 100000, 100000],
[1, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000]])
elif component == 'first_6':
q = np.array([[1, 1, 1, 100000, 100000, 100000, 100000, 100000],
[1, 1, 100000, 100000, 100000, 100000, 100000, 100000],
[1, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000]])
elif component == 'first_10':
q = np.array([[1, 1, 1, 1, 100000, 100000, 100000, 100000],
[1, 1, 1, 100000, 100000, 100000, 100000, 100000],
[1, 1, 100000, 100000, 100000, 100000, 100000, 100000],
[1, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000],
[100000, 100000, 100000, 100000, 100000, 100000, 100000, 100000]])
#q1 and q2 are some more realistic quantization values. You can try these, but no need to report any result from using 'q1' or 'q2'.
elif component == 'q1':
q = np.array([[2, 2, 2, 2, 3, 4, 5, 6],
[2, 2, 2, 2, 3, 4, 5, 6],
[2, 2, 2, 2, 4, 5, 7, 9],
[2, 2, 2, 4, 5, 7, 9, 12],
[3, 3, 4, 5, 8, 10, 12, 12],
[4, 4, 5, 7, 10, 12, 12, 12],
[5, 5, 7, 9, 12, 12, 12, 12],
[6, 6, 9, 12, 12, 12, 12, 12]])
elif component == 'q2':
q = np.array([[16, 11, 10, 16, 24, 40, 51, 61],
[12, 12, 14, 19, 26, 58, 60, 55],
[14, 13, 16, 24, 40, 57, 69, 56],
[14, 17, 22, 29, 51, 87, 80, 62],
[18, 22, 37, 56, 68, 109, 103, 77],
[24, 35, 55, 64, 81, 104, 1113, 92],
[49, 64, 78, 87, 103, 121, 120, 101],
[72, 92, 95, 98, 112, 100, 103, 99]])
else:
raise ValueError((
"Unrecognized compoents,'{comp}' was found").format(comp=component))
return q
def zigzag_points(rows, cols):
# constants for directions
UP, DOWN, RIGHT, LEFT, UP_RIGHT, DOWN_LEFT = range(6)
# move the point in different directions
def move(direction, point):
return {
UP: lambda point: (point[0] - 1, point[1]),
DOWN: lambda point: (point[0] + 1, point[1]),
LEFT: lambda point: (point[0], point[1] - 1),
RIGHT: lambda point: (point[0], point[1] + 1),
UP_RIGHT: lambda point: move(UP, move(RIGHT, point)),
DOWN_LEFT: lambda point: move(DOWN, move(LEFT, point))
}[direction](point)
# return true if point is inside the block bounds
def inbounds(point):
return 0 <= point[0] < rows and 0 <= point[1] < cols
# start in the top-left cell
point = (0, 0)
# True when moving up-right, False when moving down-left
move_up = True
for i in range(rows * cols):
yield point
if move_up:
if inbounds(move(UP_RIGHT, point)):
point = move(UP_RIGHT, point)
else:
move_up = False
if inbounds(move(RIGHT, point)):
point = move(RIGHT, point)
else:
point = move(DOWN, point)
else:
if inbounds(move(DOWN_LEFT, point)):
point = move(DOWN_LEFT, point)
else:
move_up = True
if inbounds(move(DOWN, point)):
point = move(DOWN, point)
else:
point = move(RIGHT, point)
def bits_required(n):
n = abs(n)
result = 0
while n > 0:
n >>= 1
result += 1
return result
def binstr_flip(binstr):
# check if binstr is a binary string
if not set(binstr).issubset('01'):
raise ValueError("binstr should have only '0's and '1's")
return ''.join(map(lambda c: '0' if c == '1' else '1', binstr))
def uint_to_binstr(number, size):
return bin(number)[2:][-size:].zfill(size)
def int_to_binstr(n):
if n == 0:
return ''
binstr = bin(abs(n))[2:]
# change every 0 to 1 and vice verse when n is negative
return binstr if n > 0 else binstr_flip(binstr)
def flatten(lst):
return [item for sublist in lst for item in sublist]
#Utility functions for encoding and writing to files below
def quantize(block, component):
q = load_quantization_table(component)
return (block / q).round().astype(np.int32)
def block_to_zigzag(block):
return np.array([block[point] for point in zigzag_points(*block.shape)])
def run_length_encode(arr):
# determine where the sequence is ending prematurely
last_nonzero = -1
for i, elem in enumerate(arr):
if elem != 0:
last_nonzero = i
# each symbol is a (RUNLENGTH, SIZE) tuple
symbols = []
# values are binary representations of array elements using SIZE bits
values = []
run_length = 0
for i, elem in enumerate(arr):
if i > last_nonzero:
symbols.append((0, 0))
values.append(int_to_binstr(0))
break
elif elem == 0 and run_length < 15:
run_length += 1
else:
size = bits_required(elem)
symbols.append((run_length, size))
values.append(int_to_binstr(elem))
run_length = 0
return symbols, values
def write_to_file(filepath, dc, ac, blocks_count, tables):
try:
f = open(filepath, 'w')
except FileNotFoundError as e:
raise FileNotFoundError(
"No such directory: {}".format(
os.path.dirname(filepath))) from e
#f.write('1')
for table_name in ['dc_y', 'ac_y']:
# 16 bits for 'table_size'
f.write(uint_to_binstr(len(tables[table_name]), 16))
for key, value in tables[table_name].items():
if table_name == 'dc_y':
# 4 bits for the 'category'
# 4 bits for 'code_length'
# 'code_length' bits for 'huffman_code'
f.write(uint_to_binstr(key, 4))
f.write(uint_to_binstr(len(value), 4))
f.write(value)
else:
# 4 bits for 'run_length'
# 4 bits for 'size'
# 8 bits for 'code_length'
# 'code_length' bits for 'huffman_code'
f.write(uint_to_binstr(key[0], 4))
f.write(uint_to_binstr(key[1], 4))
f.write(uint_to_binstr(len(value), 8))
f.write(value)
# 32 bits for 'blocks_count'
f.write(uint_to_binstr(blocks_count, 32))
for b in range(blocks_count):
for c in range(1):
category = bits_required(dc[b, c])
symbols, values = run_length_encode(ac[b, :, c])
dc_table = tables['dc_y']
ac_table = tables['ac_y']
f.write(dc_table[category])
f.write(int_to_binstr(dc[b, c]))
#bit_count_ac = 0
for i in range(len(symbols)):
f.write(ac_table[tuple(symbols[i])])
f.write(values[i])
#bits_written = len(ac_table[tuple(symbols[i])]) + len(values[i])
#bit_count_ac += bits_written
#print('Block {}:{} bits'.format(b,bit_count_ac))
f.close()
def HuffmanEncode_im(filepath, im):
"""Huffman encoding the gray scale image directly.
Args:
filepath : [Location to save the encoded file]
im ([numpy array]): Shape (H,W)
"""
im = im.flatten()
pixel_count = len(im)
f = open(filepath,'w')
H_Tree = HuffmanTree(im.flatten())
H_table = H_Tree.value_to_bitstring_table()
# 16 bits for 'table_size'
f.write(uint_to_binstr(len(H_table), 16))
for key, value in H_table.items():
# 4 bits for the 'category'
# 4 bits for 'code_length'
# 'code_length' bits for 'huffman_code'
f.write(uint_to_binstr(key, 4))
f.write(uint_to_binstr(len(value), 4))
f.write(value)
# 32 bits for 'pixel_count'
f.write(uint_to_binstr(pixel_count, 32))
for item in im:
f.write(H_table[item])
f.close()
def write_as_binary(string_file_path, output_file_path):
f = open(string_file_path,'r')
binary_string = f.read()
f_binary = open(output_file_path,mode='wb')
#Remainder bits are not written for simplicity.
for i in range(len(binary_string) // 8):
byte = bytes([int(binary_string[8*i:8*i+8],2)])
f_binary.write(byte)
f.close()
f_binary.close()
# Utility functions for decode
class JPEGFileReader:
TABLE_SIZE_BITS = 16
BLOCKS_COUNT_BITS = 32
DC_CODE_LENGTH_BITS = 4
CATEGORY_BITS = 4
AC_CODE_LENGTH_BITS = 8
RUN_LENGTH_BITS = 4
SIZE_BITS = 4
def __init__(self, filepath):
self.__file = open(filepath, 'r')
def read_int(self, size):
if size == 0:
return 0
# the most significant bit indicates the sign of the number
bin_num = self.__read_str(size)
if bin_num[0] == '1':
return self.__int2(bin_num)
else:
return self.__int2(binstr_flip(bin_num)) * -1
def read_dc_table(self):
table = dict()
table_size = self.__read_uint(self.TABLE_SIZE_BITS)
for _ in range(table_size):
category = self.__read_uint(self.CATEGORY_BITS)
code_length = self.__read_uint(self.DC_CODE_LENGTH_BITS)
code = self.__read_str(code_length)
table[code] = category
return table
def read_ac_table(self):
table = dict()
table_size = self.__read_uint(self.TABLE_SIZE_BITS)
for _ in range(table_size):
run_length = self.__read_uint(self.RUN_LENGTH_BITS)
size = self.__read_uint(self.SIZE_BITS)
code_length = self.__read_uint(self.AC_CODE_LENGTH_BITS)
code = self.__read_str(code_length)
table[code] = (run_length, size)
return table
def read_blocks_count(self):
return self.__read_uint(self.BLOCKS_COUNT_BITS)
def read_huffman_code(self, table):
prefix = ''
while prefix not in table:
prefix += self.__read_char()
return table[prefix]
def __read_uint(self, size):
if size <= 0:
raise ValueError("size of unsigned int should be greater than 0")
return self.__int2(self.__read_str(size))
def __read_str(self, length):
return self.__file.read(length)
def __read_char(self):
return self.__read_str(1)
def __int2(self, bin_num):
return int(bin_num, 2)
def read_image_file(filepath):
reader = JPEGFileReader(filepath)
tables = dict()
for table_name in ['dc_y', 'ac_y']:
if 'dc' in table_name:
tables[table_name] = reader.read_dc_table()
else:
tables[table_name] = reader.read_ac_table()
blocks_count = reader.read_blocks_count()
dc = np.empty((blocks_count, 1), dtype=np.int32)
ac = np.empty((blocks_count, 63, 1), dtype=np.int32)
for block_index in range(blocks_count):
for component in range(1):
dc_table = tables['dc_y']
ac_table = tables['ac_y']
category = reader.read_huffman_code(dc_table)
dc[block_index, component] = reader.read_int(category)
cells_count = 0
while cells_count < 63:
run_length, size = reader.read_huffman_code(ac_table)
if (run_length, size) == (0, 0):
while cells_count < 63:
ac[block_index, cells_count, component] = 0
cells_count += 1
else:
for i in range(run_length):
ac[block_index, cells_count, component] = 0
cells_count += 1
if size == 0:
ac[block_index, cells_count, component] = 0
else:
value = reader.read_int(size)
ac[block_index, cells_count, component] = value
cells_count += 1
return dc, ac, tables, blocks_count
def zigzag_to_block(zigzag):
# assuming that the width and the height of the block are equal
rows = cols = int(math.sqrt(len(zigzag)))
if rows * cols != len(zigzag):
raise ValueError("length of zigzag should be a perfect square")
block = np.empty((rows, cols), np.int32)
for i, point in enumerate(zigzag_points(rows, cols)):
block[point] = zigzag[i]
return block
def dequantize(block, component):
q = load_quantization_table(component)
return block * q
Encoding and Decoding
# Encoding
# Path to the input image file to be compressed.
input_file = './original.png'
# Path for the encoded code file.
output_file = './encoded_as_string'
#############################################################################
# TODO: Specify the quantization table to use here. #
# Your options: 'dc_only','first_3', 'first_6', 'first_10' #
#############################################################################
quantization_table = 'first_10'
#############################################################################
# END OF YOUR CODE #
#############################################################################
image = Image.open(input_file)
npmat = np.array(image, dtype=np.int16)[:,:,None]
rows, cols = npmat.shape[0], npmat.shape[1]
# block size: 8x8
if rows % 8 == cols % 8 == 0:
blocks_count = rows // 8 * cols // 8
else:
raise ValueError(("the width and height of the image should both be mutiples of 8"))
# dc is the top-left cell of the block, ac are all the other cells
dc = np.empty((blocks_count, 1), dtype=np.int32)
ac = np.empty((blocks_count, 63, 1), dtype=np.int32)
block_index = 0
for i in range(0, rows, 8):
for j in range(0, cols, 8):
for k in range(1):
# split 8x8 block and center the data range on zero
# [0, 255] --> [-128, 127]
block = npmat[i:i+8, j:j+8, k] - 128
dct_matrix = DCT_2D(block)
quant_matrix = quantize(dct_matrix,quantization_table)
zz = block_to_zigzag(quant_matrix)
dc[block_index, k] = zz[0]
ac[block_index, :, k] = zz[1:]
block_index += 1
H_DC_Y = HuffmanTree(np.vectorize(bits_required)(dc[:, 0]))
H_AC_Y = HuffmanTree(flatten(run_length_encode(ac[i, :, 0])[0] for i in range(blocks_count)))
tables = {'dc_y': H_DC_Y.value_to_bitstring_table(), 'ac_y': H_AC_Y.value_to_bitstring_table()}
write_to_file(output_file, dc, ac, blocks_count, tables)
write_as_binary(output_file, 'encoded_as_binary')
print('Finished encoding and writing data to file.')
# Printing the original image size and compressed file size
file_name = "original.png"
file_stats = os.stat(file_name)
size_ori = file_stats.st_size / (1024)
print('Orignal image size is {:.0f} kB'.format(size_ori))
file_name = "encoded_as_binary"
file_stats = os.stat(file_name)
size_compressed = file_stats.st_size / (1024)
print('Compressed file size is {:.0f} kB'.format(size_compressed))
print('Compression ratio is {:.2f}'.format(size_compressed/size_ori))
# Decoding
# Path to the encoded as string file.
encoded_string_file = 'encoded_as_string'
dc, ac, tables, blocks_count = read_image_file(encoded_string_file)
# Assuming that the block is a 8x8 square
block_side = 8
# Assuming that the image height and width are equal
image_side = int(math.sqrt(blocks_count)) * block_side
blocks_per_line = image_side // block_side
npmat = np.empty((image_side, image_side, 1), dtype=np.uint8)
for block_index in range(blocks_count):
i = block_index // blocks_per_line * block_side
j = block_index % blocks_per_line * block_side
for c in range(1):
zigzag = [dc[block_index, c]] + list(ac[block_index, :, c])
quant_matrix = zigzag_to_block(zigzag)
dct_matrix = dequantize(quant_matrix, quantization_table)
block = IDCT_2D(dct_matrix)
npmat[i:i+8, j:j+8, c] = (block + 128).clip(0,255)
print('Finished decoding file.')
plt.figure(figsize=(8,8))
plt.imshow(npmat[:,:,0],cmap='gray')
plt.title('Decoded_image')
image = Image.fromarray(npmat[:,:,0], 'L')
image.save('Decoded.png', compress_level=0)
Finished encoding and writing data to file.
Orignal image size is 226 kB
Compressed file size is 36 kB
Compression ratio is 0.16
Finished decoding file.

# Encoding
# Path to the input image file to be compressed.
input_file = './original.png'
# Path for the encoded code file.
output_file = './encoded_as_string'
#############################################################################
# TODO: Specify the quantization table to use here. #
# Your options: 'dc_only','first_3', 'first_6', 'first_10' #
#############################################################################
quantization_table = 'first_6'
#############################################################################
# END OF YOUR CODE #
#############################################################################
image = Image.open(input_file)
npmat = np.array(image, dtype=np.int16)[:,:,None]
rows, cols = npmat.shape[0], npmat.shape[1]
# block size: 8x8
if rows % 8 == cols % 8 == 0:
blocks_count = rows // 8 * cols // 8
else:
raise ValueError(("the width and height of the image should both be mutiples of 8"))
# dc is the top-left cell of the block, ac are all the other cells
dc = np.empty((blocks_count, 1), dtype=np.int32)
ac = np.empty((blocks_count, 63, 1), dtype=np.int32)
block_index = 0
for i in range(0, rows, 8):
for j in range(0, cols, 8):
for k in range(1):
# split 8x8 block and center the data range on zero
# [0, 255] --> [-128, 127]
block = npmat[i:i+8, j:j+8, k] - 128
dct_matrix = DCT_2D(block)
quant_matrix = quantize(dct_matrix,quantization_table)
zz = block_to_zigzag(quant_matrix)
dc[block_index, k] = zz[0]
ac[block_index, :, k] = zz[1:]
block_index += 1
H_DC_Y = HuffmanTree(np.vectorize(bits_required)(dc[:, 0]))
H_AC_Y = HuffmanTree(flatten(run_length_encode(ac[i, :, 0])[0] for i in range(blocks_count)))
tables = {'dc_y': H_DC_Y.value_to_bitstring_table(), 'ac_y': H_AC_Y.value_to_bitstring_table()}
write_to_file(output_file, dc, ac, blocks_count, tables)
write_as_binary(output_file, 'encoded_as_binary')
print('Finished encoding and writing data to file.')
# Printing the original image size and compressed file size
file_name = "original.png"
file_stats = os.stat(file_name)
size_ori = file_stats.st_size / (1024)
print('Orignal image size is {:.0f} kB'.format(size_ori))
file_name = "encoded_as_binary"
file_stats = os.stat(file_name)
size_compressed = file_stats.st_size / (1024)
print('Compressed file size is {:.0f} kB'.format(size_compressed))
print('Compression ratio is {:.2f}'.format(size_compressed/size_ori))
# Decoding
# Path to the encoded as string file.
encoded_string_file = 'encoded_as_string'
dc, ac, tables, blocks_count = read_image_file(encoded_string_file)
# Assuming that the block is a 8x8 square
block_side = 8
# Assuming that the image height and width are equal
image_side = int(math.sqrt(blocks_count)) * block_side
blocks_per_line = image_side // block_side
npmat = np.empty((image_side, image_side, 1), dtype=np.uint8)
for block_index in range(blocks_count):
i = block_index // blocks_per_line * block_side
j = block_index % blocks_per_line * block_side
for c in range(1):
zigzag = [dc[block_index, c]] + list(ac[block_index, :, c])
quant_matrix = zigzag_to_block(zigzag)
dct_matrix = dequantize(quant_matrix, quantization_table)
block = IDCT_2D(dct_matrix)
npmat[i:i+8, j:j+8, c] = (block + 128).clip(0,255)
print('Finished decoding file.')
plt.figure(figsize=(8,8))
plt.imshow(npmat[:,:,0],cmap='gray')
plt.title('Decoded_image')
image = Image.fromarray(npmat[:,:,0], 'L')
image.save('Decoded.png', compress_level=0)
Finished encoding and writing data to file.
Orignal image size is 226 kB
Compressed file size is 23 kB
Compression ratio is 0.10
Finished decoding file.

# Encoding
# Path to the input image file to be compressed.
input_file = './original.png'
# Path for the encoded code file.
output_file = './encoded_as_string'
#############################################################################
# TODO: Specify the quantization table to use here. #
# Your options: 'dc_only','first_3', 'first_6', 'first_10' #
#############################################################################
quantization_table = 'first_3'
#############################################################################
# END OF YOUR CODE #
#############################################################################
image = Image.open(input_file)
npmat = np.array(image, dtype=np.int16)[:,:,None]
rows, cols = npmat.shape[0], npmat.shape[1]
# block size: 8x8
if rows % 8 == cols % 8 == 0:
blocks_count = rows // 8 * cols // 8
else:
raise ValueError(("the width and height of the image should both be mutiples of 8"))
# dc is the top-left cell of the block, ac are all the other cells
dc = np.empty((blocks_count, 1), dtype=np.int32)
ac = np.empty((blocks_count, 63, 1), dtype=np.int32)
block_index = 0
for i in range(0, rows, 8):
for j in range(0, cols, 8):
for k in range(1):
# split 8x8 block and center the data range on zero
# [0, 255] --> [-128, 127]
block = npmat[i:i+8, j:j+8, k] - 128
dct_matrix = DCT_2D(block)
quant_matrix = quantize(dct_matrix,quantization_table)
zz = block_to_zigzag(quant_matrix)
dc[block_index, k] = zz[0]
ac[block_index, :, k] = zz[1:]
block_index += 1
H_DC_Y = HuffmanTree(np.vectorize(bits_required)(dc[:, 0]))
H_AC_Y = HuffmanTree(flatten(run_length_encode(ac[i, :, 0])[0] for i in range(blocks_count)))
tables = {'dc_y': H_DC_Y.value_to_bitstring_table(), 'ac_y': H_AC_Y.value_to_bitstring_table()}
write_to_file(output_file, dc, ac, blocks_count, tables)
write_as_binary(output_file, 'encoded_as_binary')
print('Finished encoding and writing data to file.')
# Printing the original image size and compressed file size
file_name = "original.png"
file_stats = os.stat(file_name)
size_ori = file_stats.st_size / (1024)
print('Orignal image size is {:.0f} kB'.format(size_ori))
file_name = "encoded_as_binary"
file_stats = os.stat(file_name)
size_compressed = file_stats.st_size / (1024)
print('Compressed file size is {:.0f} kB'.format(size_compressed))
print('Compression ratio is {:.2f}'.format(size_compressed/size_ori))
# Decoding
# Path to the encoded as string file.
encoded_string_file = 'encoded_as_string'
dc, ac, tables, blocks_count = read_image_file(encoded_string_file)
# Assuming that the block is a 8x8 square
block_side = 8
# Assuming that the image height and width are equal
image_side = int(math.sqrt(blocks_count)) * block_side
blocks_per_line = image_side // block_side
npmat = np.empty((image_side, image_side, 1), dtype=np.uint8)
for block_index in range(blocks_count):
i = block_index // blocks_per_line * block_side
j = block_index % blocks_per_line * block_side
for c in range(1):
zigzag = [dc[block_index, c]] + list(ac[block_index, :, c])
quant_matrix = zigzag_to_block(zigzag)
dct_matrix = dequantize(quant_matrix, quantization_table)
block = IDCT_2D(dct_matrix)
npmat[i:i+8, j:j+8, c] = (block + 128).clip(0,255)
print('Finished decoding file.')
plt.figure(figsize=(8,8))
plt.imshow(npmat[:,:,0],cmap='gray')
plt.title('Decoded_image')
image = Image.fromarray(npmat[:,:,0], 'L')
image.save('Decoded.png', compress_level=0)
Finished encoding and writing data to file.
Orignal image size is 226 kB
Compressed file size is 13 kB
Compression ratio is 0.06
Finished decoding file.

# Encoding
# Path to the input image file to be compressed.
input_file = './original.png'
# Path for the encoded code file.
output_file = './encoded_as_string'
#############################################################################
# TODO: Specify the quantization table to use here. #
# Your options: 'dc_only','first_3', 'first_6', 'first_10' #
#############################################################################
quantization_table = 'dc_only'
#############################################################################
# END OF YOUR CODE #
#############################################################################
image = Image.open(input_file)
npmat = np.array(image, dtype=np.int16)[:,:,None]
rows, cols = npmat.shape[0], npmat.shape[1]
# block size: 8x8
if rows % 8 == cols % 8 == 0:
blocks_count = rows // 8 * cols // 8
else:
raise ValueError(("the width and height of the image should both be mutiples of 8"))
# dc is the top-left cell of the block, ac are all the other cells
dc = np.empty((blocks_count, 1), dtype=np.int32)
ac = np.empty((blocks_count, 63, 1), dtype=np.int32)
block_index = 0
for i in range(0, rows, 8):
for j in range(0, cols, 8):
for k in range(1):
# split 8x8 block and center the data range on zero
# [0, 255] --> [-128, 127]
block = npmat[i:i+8, j:j+8, k] - 128
dct_matrix = DCT_2D(block)
quant_matrix = quantize(dct_matrix,quantization_table)
zz = block_to_zigzag(quant_matrix)
dc[block_index, k] = zz[0]
ac[block_index, :, k] = zz[1:]
block_index += 1
H_DC_Y = HuffmanTree(np.vectorize(bits_required)(dc[:, 0]))
H_AC_Y = HuffmanTree(flatten(run_length_encode(ac[i, :, 0])[0] for i in range(blocks_count)))
tables = {'dc_y': H_DC_Y.value_to_bitstring_table(), 'ac_y': H_AC_Y.value_to_bitstring_table()}
write_to_file(output_file, dc, ac, blocks_count, tables)
write_as_binary(output_file, 'encoded_as_binary')
print('Finished encoding and writing data to file.')
# Printing the original image size and compressed file size
file_name = "original.png"
file_stats = os.stat(file_name)
size_ori = file_stats.st_size / (1024)
print('Orignal image size is {:.0f} kB'.format(size_ori))
file_name = "encoded_as_binary"
file_stats = os.stat(file_name)
size_compressed = file_stats.st_size / (1024)
print('Compressed file size is {:.0f} kB'.format(size_compressed))
print('Compression ratio is {:.2f}'.format(size_compressed/size_ori))
# Decoding
# Path to the encoded as string file.
encoded_string_file = 'encoded_as_string'
dc, ac, tables, blocks_count = read_image_file(encoded_string_file)
# Assuming that the block is a 8x8 square
block_side = 8
# Assuming that the image height and width are equal
image_side = int(math.sqrt(blocks_count)) * block_side
blocks_per_line = image_side // block_side
npmat = np.empty((image_side, image_side, 1), dtype=np.uint8)
for block_index in range(blocks_count):
i = block_index // blocks_per_line * block_side
j = block_index % blocks_per_line * block_side
for c in range(1):
zigzag = [dc[block_index, c]] + list(ac[block_index, :, c])
quant_matrix = zigzag_to_block(zigzag)
dct_matrix = dequantize(quant_matrix, quantization_table)
block = IDCT_2D(dct_matrix)
npmat[i:i+8, j:j+8, c] = (block + 128).clip(0,255)
print('Finished decoding file.')
plt.figure(figsize=(8,8))
plt.imshow(npmat[:,:,0],cmap='gray')
plt.title('Decoded_image')
image = Image.fromarray(npmat[:,:,0], 'L')
image.save('Decoded.png', compress_level=0)
Finished encoding and writing data to file.
Orignal image size is 226 kB
Compressed file size is 5 kB
Compression ratio is 0.02
Finished decoding file.

