2 - 3) EDA - Vital and Laboratory variables
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as sns
import missingno as msno
from utils import my_histogram, make_stacked_table, my_stacked_barplot
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
import warnings
warnings.filterwarnings('ignore')
pd.set_option('display.max_columns', None)
df_train = pd.read_csv("../data/df_train.csv")
df_test = pd.read_csv("../data/df_test.csv")
- There are 8 vital signs variables and 26 laboratory variables.
- Vital signs
- HR: Heart rate (beats per minute)
- O2Sat: Pulse oximetry (%)
- Temp: Temperature (Deg C)
- SBP: Systolic BP (mm Hg)
- MAP: Mean arterial pressure (mm Hg)
- DBP: Diastolic BP (mm Hg)
- Resp: Respiration rate (breaths per minute)
- EtCO2: End tidal carbon dioxide (mm Hg)
- Laboratory values
- BaseExcess: Measure of excess bicarbonate (mmol/L)
- HCO3: Bicarbonate (mmol/L)
- FiO2: Fraction of inspired oxygen (%)
- pH: N/A
- PaCO2: Partial pressure of carbon dioxide from arterial blood (mm Hg)
- SaO2: Oxygen saturation from arterial blood (%)
- AST: Aspartate transaminase (IU/L)
- BUN: Blood urea nitrogen (mg/dL)
- Alkalinephos: Alkaline phosphatase (IU/L)
- Calcium: (mg/dL)
- Chloride: (mmol/L)
- Creatinine: (mg/dL)
- Bilirubin_direct: Bilirubin direct (mg/dL)
- Glucose: Serum glucose (mg/dL)
- Lactate: Lactic acid (mg/dL)
- Magnesium: (mmol/dL)
- Phosphate: (mg/dL)
- Potassium: (mmol/L)
- Bilirubin_total: Total bilirubin (mg/dL)
- TroponinI: Troponin I (ng/mL)
- Hct: Hematocrit (%)
- Hgb: Hemoglobin (g/dL)
- PTT: partial thromboplastin time (seconds)
- WBC: Leukocyte count (count*10^3/µL)
- Fibrinogen: (mg/dL)
- Platelets: (count*10^3/µL)
- Vital signs
1. Shock index & BUN/CR
- There are some researches that follwing features are important in predicting the sepsis (Henry, K. E., Hager, D. N., Pronovost, P. J., and Saria, S. A targeted real-time early warning score (trewscore) for septic shock. Science Translational Medicine 7, 299 (2015), 299ra122–299ra122.):
- ShockIndex(t) = $\frac{\text{HR(t)}}{SBP(t)}$
- BUN/CR(t) = $\frac{\text{BUN(t)}}{Creatinine(t)}$
- Let’s make these features.
df_train["ShockIndex"] = df_train.HR / df_train.SBP
df_test["ShockIndex"] = df_test.HR / df_test.SBP
df_train["BUN/CR"] = df_train.BUN / df_train.Creatinine
df_test["BUN/CR"] = df_test.BUN / df_test.Creatinine
2. Measurement time
- Let’s incorporate the measurment time information of each variables:
- Beginning time: Time of the first measurment
- End time: Time of the last measurment
- Frequency: Ratio between the total number of the measurment and the maximum hour of the patient
target_col_list = ['BUN', 'Creatinine', 'DBP', 'HR', 'Hct', 'Hgb',
'MAP', 'Magnesium', 'O2Sat', 'Phosphate', 'Platelets',
'Potassium', 'Resp', 'SBP', 'Temp', 'WBC', 'BaseExcess', 'Chloride',
'FiO2', 'Glucose', 'HCO3', 'PaCO2', 'SaO2', 'pH', 'Calcium',
'Fibrinogen', 'Lactate', 'PTT', 'TroponinI', 'AST', 'Alkalinephos',
'Bilirubin_total', 'EtCO2', 'Bilirubin_direct', 'ShockIndex','BUN/CR']
df_train_measurment_time_info = df_train[["ID"]].drop_duplicates()
df_test_measurment_time_info = df_test[["ID"]].drop_duplicates()
for target_col in target_col_list:
begin_time = df_train[~df_train[target_col].isna()].groupby("ID").Hour.min().reset_index()
end_time = df_train[~df_train[target_col].isna()].groupby("ID").Hour.max().reset_index()
freq = (df_train[~df_train[target_col].isna()].groupby("ID").Hour.count() / df_train.groupby("ID").Hour.count()).reset_index()
df_train_measurment_time_info = df_train_measurment_time_info.merge(begin_time.rename(columns = {"Hour": f"{target_col}_beg_time"}), how = "left", on = "ID") \
.merge(end_time.rename(columns = {"Hour": f"{target_col}_end_time"}), how = "left", on = "ID") \
.merge(freq.rename(columns = {"Hour": f"{target_col}_freq"}), how = "left", on = "ID")
for target_col in target_col_list:
begin_time = df_test[~df_test[target_col].isna()].groupby("ID").Hour.min().reset_index()
end_time = df_test[~df_test[target_col].isna()].groupby("ID").Hour.max().reset_index()
freq = (df_test[~df_test[target_col].isna()].groupby("ID").Hour.count() / df_test.groupby("ID").Hour.count()).reset_index()
df_test_measurment_time_info = df_test_measurment_time_info.merge(begin_time.rename(columns = {"Hour": f"{target_col}_beg_time"}), how = "left", on = "ID") \
.merge(end_time.rename(columns = {"Hour": f"{target_col}_end_time"}), how = "left", on = "ID") \
.merge(freq.rename(columns = {"Hour": f"{target_col}_freq"}), how = "left", on = "ID")
- Also merge the information that shows how long each patient has stayed (the maximum hour of each patient).
df_train_measurment_time_info = df_train_measurment_time_info.merge(df_train.groupby("ID").Hour.count().reset_index().rename(columns = {"Hour": "length_of_stay"}),
how = "left", on = "ID")
df_test_measurment_time_info = df_test_measurment_time_info.merge(df_test.groupby("ID").Hour.count().reset_index().rename(columns = {"Hour": "length_of_stay"}),
how = "left", on = "ID")
df_train_measurment_time_info
| ID | BUN_beg_time | BUN_end_time | BUN_freq | Creatinine_beg_time | Creatinine_end_time | Creatinine_freq | DBP_beg_time | DBP_end_time | DBP_freq | HR_beg_time | HR_end_time | HR_freq | Hct_beg_time | Hct_end_time | Hct_freq | Hgb_beg_time | Hgb_end_time | Hgb_freq | MAP_beg_time | MAP_end_time | MAP_freq | Magnesium_beg_time | Magnesium_end_time | Magnesium_freq | O2Sat_beg_time | O2Sat_end_time | O2Sat_freq | Phosphate_beg_time | Phosphate_end_time | Phosphate_freq | Platelets_beg_time | Platelets_end_time | Platelets_freq | Potassium_beg_time | Potassium_end_time | Potassium_freq | Resp_beg_time | Resp_end_time | Resp_freq | SBP_beg_time | SBP_end_time | SBP_freq | Temp_beg_time | Temp_end_time | Temp_freq | WBC_beg_time | WBC_end_time | WBC_freq | BaseExcess_beg_time | BaseExcess_end_time | BaseExcess_freq | Chloride_beg_time | Chloride_end_time | Chloride_freq | FiO2_beg_time | FiO2_end_time | FiO2_freq | Glucose_beg_time | Glucose_end_time | Glucose_freq | HCO3_beg_time | HCO3_end_time | HCO3_freq | PaCO2_beg_time | PaCO2_end_time | PaCO2_freq | SaO2_beg_time | SaO2_end_time | SaO2_freq | pH_beg_time | pH_end_time | pH_freq | Calcium_beg_time | Calcium_end_time | Calcium_freq | Fibrinogen_beg_time | Fibrinogen_end_time | Fibrinogen_freq | Lactate_beg_time | Lactate_end_time | Lactate_freq | PTT_beg_time | PTT_end_time | PTT_freq | TroponinI_beg_time | TroponinI_end_time | TroponinI_freq | AST_beg_time | AST_end_time | AST_freq | Alkalinephos_beg_time | Alkalinephos_end_time | Alkalinephos_freq | Bilirubin_total_beg_time | Bilirubin_total_end_time | Bilirubin_total_freq | EtCO2_beg_time | EtCO2_end_time | EtCO2_freq | Bilirubin_direct_beg_time | Bilirubin_direct_end_time | Bilirubin_direct_freq | ShockIndex_beg_time | ShockIndex_end_time | ShockIndex_freq | BUN/CR_beg_time | BUN/CR_end_time | BUN/CR_freq | length_of_stay | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 17.0 | 18.0 | 0.090909 | 17.0 | 17.0 | 0.045455 | 4.0 | 24.0 | 0.909091 | 4.0 | 24.0 | 0.909091 | 11.0 | 18.0 | 0.181818 | 17.0 | 18.0 | 0.090909 | 4.0 | 24.0 | 0.909091 | NaN | NaN | NaN | 4.0 | 24.0 | 0.909091 | NaN | NaN | NaN | NaN | NaN | NaN | 17.0 | 18.0 | 0.090909 | 4.0 | 24.0 | 0.909091 | 4.0 | 24.0 | 0.863636 | 5.0 | 23.0 | 0.409091 | 17.0 | 18.0 | 0.090909 | 4.0 | 5.0 | 0.090909 | 17.0 | 18.0 | 0.090909 | 4.0 | 5.0 | 0.090909 | 4.0 | 17.0 | 0.090909 | 17.0 | 18.0 | 0.090909 | 4.0 | 4.0 | 0.045455 | 4.0 | 5.0 | 0.090909 | 4.0 | 5.0 | 0.090909 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 4.0 | 24.0 | 0.863636 | 17.0 | 17.0 | 0.045455 | 22 |
| 1 | 3 | 15.0 | 38.0 | 0.051724 | 15.0 | 38.0 | 0.051724 | 2.0 | 41.0 | 0.568966 | 2.0 | 58.0 | 0.862069 | 15.0 | 38.0 | 0.137931 | 15.0 | 38.0 | 0.137931 | 2.0 | 58.0 | 0.896552 | 8.0 | 48.0 | 0.086207 | 2.0 | 58.0 | 0.620690 | 15.0 | 15.0 | 0.017241 | 15.0 | 38.0 | 0.086207 | 8.0 | 48.0 | 0.172414 | 2.0 | 58.0 | 0.879310 | 2.0 | 58.0 | 0.879310 | 4.0 | 56.0 | 0.500000 | 15.0 | 38.0 | 0.068966 | 9.0 | 40.0 | 0.189655 | 15.0 | 38.0 | 0.051724 | 22.0 | 25.0 | 0.051724 | 8.0 | 48.0 | 0.189655 | 15.0 | 22.0 | 0.034483 | 9.0 | 40.0 | 0.189655 | 22.0 | 40.0 | 0.120690 | 9.0 | 40.0 | 0.189655 | 8.0 | 48.0 | 0.051724 | 15.0 | 21.0 | 0.034483 | 9.0 | 25.0 | 0.086207 | 15.0 | 38.0 | 0.068966 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 58.0 | 0.827586 | 15.0 | 38.0 | 0.051724 | 58 |
| 2 | 4 | NaN | NaN | NaN | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.947368 | 2.0 | 38.0 | 0.868421 | 16.0 | 16.0 | 0.026316 | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.921053 | NaN | NaN | NaN | 2.0 | 38.0 | 0.894737 | NaN | NaN | NaN | NaN | NaN | NaN | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.947368 | 2.0 | 38.0 | 0.868421 | 3.0 | 35.0 | 0.184211 | 16.0 | 16.0 | 0.026316 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 4.0 | 32.0 | 0.184211 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 16.0 | 16.0 | 0.026316 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 27.0 | 27.0 | 0.026316 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 38.0 | 0.789474 | NaN | NaN | NaN | 38 |
| 3 | 5 | 6.0 | 124.0 | 0.044776 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.902985 | 2.0 | 134.0 | 0.940299 | 6.0 | 124.0 | 0.044776 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.955224 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.880597 | 6.0 | 124.0 | 0.037313 | 31.0 | 124.0 | 0.029851 | 6.0 | 124.0 | 0.067164 | 3.0 | 134.0 | 0.917910 | 2.0 | 133.0 | 0.917910 | 2.0 | 134.0 | 0.231343 | 6.0 | 124.0 | 0.044776 | NaN | NaN | NaN | NaN | NaN | NaN | 48.0 | 133.0 | 0.097015 | 6.0 | 130.0 | 0.186567 | NaN | NaN | NaN | 39.0 | 133.0 | 0.104478 | 31.0 | 133.0 | 0.104478 | 31.0 | 133.0 | 0.104478 | 6.0 | 124.0 | 0.059701 | NaN | NaN | NaN | 72.0 | 83.0 | 0.022388 | 6.0 | 6.0 | 0.007463 | 6.0 | 86.0 | 0.014925 | 6.0 | 6.0 | 0.007463 | 6.0 | 6.0 | 0.007463 | 6.0 | 6.0 | 0.007463 | 46.0 | 134.0 | 0.641791 | NaN | NaN | NaN | 2.0 | 133.0 | 0.880597 | 6.0 | 124.0 | 0.044776 | 134 |
| 4 | 6 | 3.0 | 28.0 | 0.039216 | 3.0 | 28.0 | 0.039216 | NaN | NaN | NaN | 2.0 | 50.0 | 0.803922 | 3.0 | 22.0 | 0.058824 | 3.0 | 28.0 | 0.058824 | 2.0 | 49.0 | 0.803922 | 3.0 | 28.0 | 0.039216 | 2.0 | 50.0 | 0.764706 | 3.0 | 28.0 | 0.039216 | 3.0 | 28.0 | 0.058824 | 3.0 | 28.0 | 0.058824 | 2.0 | 50.0 | 0.784314 | 2.0 | 49.0 | 0.784314 | 4.0 | 47.0 | 0.215686 | 3.0 | 28.0 | 0.058824 | NaN | NaN | NaN | 3.0 | 28.0 | 0.039216 | NaN | NaN | NaN | 3.0 | 28.0 | 0.039216 | 3.0 | 28.0 | 0.039216 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.0 | 28.0 | 0.039216 | 3.0 | 3.0 | 0.019608 | NaN | NaN | NaN | 3.0 | 28.0 | 0.058824 | NaN | NaN | NaN | 22.0 | 28.0 | 0.039216 | 28.0 | 28.0 | 0.019608 | 22.0 | 28.0 | 0.039216 | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 49.0 | 0.745098 | 3.0 | 28.0 | 0.039216 | 51 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15139 | 21628 | 17.0 | 17.0 | 0.035714 | 17.0 | 17.0 | 0.035714 | 10.0 | 21.0 | 0.428571 | 10.0 | 36.0 | 0.964286 | 10.0 | 17.0 | 0.071429 | 17.0 | 17.0 | 0.035714 | 10.0 | 36.0 | 0.857143 | NaN | NaN | NaN | 10.0 | 36.0 | 0.892857 | NaN | NaN | NaN | 17.0 | 17.0 | 0.035714 | 10.0 | 17.0 | 0.071429 | 10.0 | 36.0 | 0.928571 | 10.0 | 20.0 | 0.392857 | 10.0 | 35.0 | 0.250000 | 17.0 | 17.0 | 0.035714 | NaN | NaN | NaN | 10.0 | 17.0 | 0.071429 | NaN | NaN | NaN | 17.0 | 17.0 | 0.035714 | 17.0 | 17.0 | 0.035714 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 17.0 | 17.0 | 0.035714 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 10.0 | 20.0 | 0.392857 | 17.0 | 17.0 | 0.035714 | 28 |
| 15140 | 21629 | 15.0 | 39.0 | 0.035714 | 15.0 | 39.0 | 0.035714 | 2.0 | 56.0 | 0.857143 | 2.0 | 56.0 | 0.839286 | 15.0 | 39.0 | 0.053571 | 15.0 | 39.0 | 0.053571 | 2.0 | 56.0 | 0.821429 | 15.0 | 15.0 | 0.017857 | 2.0 | 56.0 | 0.821429 | NaN | NaN | NaN | 15.0 | 39.0 | 0.035714 | 3.0 | 39.0 | 0.089286 | 2.0 | 56.0 | 0.839286 | 2.0 | 56.0 | 0.803571 | 2.0 | 56.0 | 0.482143 | 15.0 | 39.0 | 0.035714 | NaN | NaN | NaN | NaN | NaN | NaN | 3.0 | 9.0 | 0.053571 | 2.0 | 44.0 | 0.410714 | NaN | NaN | NaN | 3.0 | 9.0 | 0.071429 | 3.0 | 9.0 | 0.071429 | 3.0 | 9.0 | 0.053571 | 3.0 | 39.0 | 0.107143 | NaN | NaN | NaN | 3.0 | 9.0 | 0.053571 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 56.0 | 0.767857 | 15.0 | 39.0 | 0.035714 | 56 |
| 15141 | 21630 | 13.0 | 44.0 | 0.069767 | 13.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.930233 | 4.0 | 45.0 | 0.930233 | 5.0 | 44.0 | 0.093023 | 18.0 | 44.0 | 0.046512 | 4.0 | 45.0 | 0.930233 | 5.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.883721 | 13.0 | 44.0 | 0.069767 | 18.0 | 44.0 | 0.046512 | 13.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.767442 | 4.0 | 45.0 | 0.930233 | 4.0 | 45.0 | 0.906977 | 18.0 | 44.0 | 0.046512 | 4.0 | 43.0 | 0.139535 | 18.0 | 44.0 | 0.046512 | NaN | NaN | NaN | 4.0 | 44.0 | 0.162791 | 13.0 | 44.0 | 0.069767 | 4.0 | 43.0 | 0.116279 | 45.0 | 45.0 | 0.023256 | 4.0 | 43.0 | 0.139535 | 13.0 | 18.0 | 0.046512 | NaN | NaN | NaN | 13.0 | 18.0 | 0.046512 | 18.0 | 18.0 | 0.023256 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 4.0 | 45.0 | 0.906977 | 13.0 | 44.0 | 0.069767 | 43 |
| 15142 | 21632 | 5.0 | 9.0 | 0.142857 | 5.0 | 5.0 | 0.071429 | 2.0 | 14.0 | 0.857143 | 2.0 | 14.0 | 0.928571 | 5.0 | 13.0 | 0.285714 | 5.0 | 13.0 | 0.285714 | 2.0 | 14.0 | 0.928571 | 5.0 | 9.0 | 0.142857 | 2.0 | 14.0 | 0.857143 | 5.0 | 9.0 | 0.142857 | 5.0 | 9.0 | 0.142857 | 5.0 | 9.0 | 0.214286 | 2.0 | 14.0 | 0.928571 | 2.0 | 14.0 | 0.785714 | 4.0 | 14.0 | 0.285714 | 5.0 | 9.0 | 0.142857 | 5.0 | 13.0 | 0.285714 | 5.0 | 9.0 | 0.142857 | 5.0 | 14.0 | 0.285714 | 9.0 | 9.0 | 0.071429 | 5.0 | 9.0 | 0.142857 | 5.0 | 10.0 | 0.214286 | 5.0 | 13.0 | 0.285714 | 5.0 | 13.0 | 0.285714 | 5.0 | 9.0 | 0.142857 | NaN | NaN | NaN | 5.0 | 8.0 | 0.142857 | 5.0 | 5.0 | 0.071429 | NaN | NaN | NaN | 9.0 | 9.0 | 0.071429 | 9.0 | 9.0 | 0.071429 | 9.0 | 9.0 | 0.071429 | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 14.0 | 0.785714 | 5.0 | 5.0 | 0.071429 | 14 |
| 15143 | 21633 | 2.0 | 6.0 | 0.157895 | 5.0 | 5.0 | 0.052632 | NaN | NaN | NaN | 2.0 | 19.0 | 0.894737 | 2.0 | 19.0 | 0.368421 | 5.0 | 6.0 | 0.105263 | 2.0 | 18.0 | 0.842105 | 2.0 | 6.0 | 0.157895 | 2.0 | 19.0 | 0.789474 | 5.0 | 5.0 | 0.052632 | 5.0 | 5.0 | 0.052632 | 2.0 | 6.0 | 0.157895 | 2.0 | 19.0 | 0.894737 | 2.0 | 19.0 | 0.894737 | 3.0 | 16.0 | 0.263158 | 5.0 | 6.0 | 0.105263 | NaN | NaN | NaN | 2.0 | 6.0 | 0.157895 | NaN | NaN | NaN | 5.0 | 5.0 | 0.052632 | 2.0 | 6.0 | 0.157895 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 5.0 | 5.0 | 0.052632 | NaN | NaN | NaN | NaN | NaN | NaN | 5.0 | 5.0 | 0.052632 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2.0 | 19.0 | 0.842105 | 5.0 | 5.0 | 0.052632 | 19 |
15144 rows × 110 columns
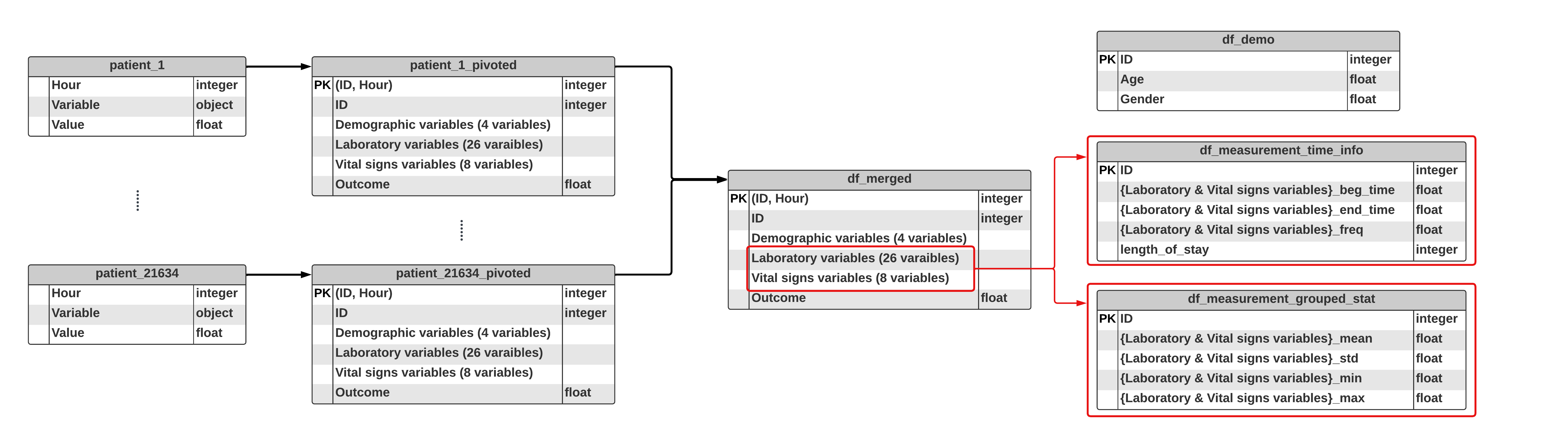
msno.matrix(df_train_measurment_time_info)
<AxesSubplot:>
- Drop the variables with more than 20% missing values.
target_col = df_train_measurment_time_info.isna().sum()[(df_train_measurment_time_info.isna().sum() / df_train_measurment_time_info.shape[0]) < 0.2].reset_index()["index"]
df_train_measurment_time_info = df_train_measurment_time_info[target_col]
df_test_measurment_time_info = df_test_measurment_time_info[target_col]
df_train_measurment_time_info
| ID | BUN_beg_time | BUN_end_time | BUN_freq | Creatinine_beg_time | Creatinine_end_time | Creatinine_freq | DBP_beg_time | DBP_end_time | DBP_freq | HR_beg_time | HR_end_time | HR_freq | Hct_beg_time | Hct_end_time | Hct_freq | Hgb_beg_time | Hgb_end_time | Hgb_freq | MAP_beg_time | MAP_end_time | MAP_freq | Magnesium_beg_time | Magnesium_end_time | Magnesium_freq | O2Sat_beg_time | O2Sat_end_time | O2Sat_freq | Platelets_beg_time | Platelets_end_time | Platelets_freq | Potassium_beg_time | Potassium_end_time | Potassium_freq | Resp_beg_time | Resp_end_time | Resp_freq | SBP_beg_time | SBP_end_time | SBP_freq | Temp_beg_time | Temp_end_time | Temp_freq | WBC_beg_time | WBC_end_time | WBC_freq | Glucose_beg_time | Glucose_end_time | Glucose_freq | Calcium_beg_time | Calcium_end_time | Calcium_freq | ShockIndex_beg_time | ShockIndex_end_time | ShockIndex_freq | BUN/CR_beg_time | BUN/CR_end_time | BUN/CR_freq | length_of_stay | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 17.0 | 18.0 | 0.090909 | 17.0 | 17.0 | 0.045455 | 4.0 | 24.0 | 0.909091 | 4.0 | 24.0 | 0.909091 | 11.0 | 18.0 | 0.181818 | 17.0 | 18.0 | 0.090909 | 4.0 | 24.0 | 0.909091 | NaN | NaN | NaN | 4.0 | 24.0 | 0.909091 | NaN | NaN | NaN | 17.0 | 18.0 | 0.090909 | 4.0 | 24.0 | 0.909091 | 4.0 | 24.0 | 0.863636 | 5.0 | 23.0 | 0.409091 | 17.0 | 18.0 | 0.090909 | 4.0 | 17.0 | 0.090909 | NaN | NaN | NaN | 4.0 | 24.0 | 0.863636 | 17.0 | 17.0 | 0.045455 | 22 |
| 1 | 3 | 15.0 | 38.0 | 0.051724 | 15.0 | 38.0 | 0.051724 | 2.0 | 41.0 | 0.568966 | 2.0 | 58.0 | 0.862069 | 15.0 | 38.0 | 0.137931 | 15.0 | 38.0 | 0.137931 | 2.0 | 58.0 | 0.896552 | 8.0 | 48.0 | 0.086207 | 2.0 | 58.0 | 0.620690 | 15.0 | 38.0 | 0.086207 | 8.0 | 48.0 | 0.172414 | 2.0 | 58.0 | 0.879310 | 2.0 | 58.0 | 0.879310 | 4.0 | 56.0 | 0.500000 | 15.0 | 38.0 | 0.068966 | 8.0 | 48.0 | 0.189655 | 8.0 | 48.0 | 0.051724 | 2.0 | 58.0 | 0.827586 | 15.0 | 38.0 | 0.051724 | 58 |
| 2 | 4 | NaN | NaN | NaN | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.947368 | 2.0 | 38.0 | 0.868421 | 16.0 | 16.0 | 0.026316 | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.921053 | NaN | NaN | NaN | 2.0 | 38.0 | 0.894737 | NaN | NaN | NaN | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.947368 | 2.0 | 38.0 | 0.868421 | 3.0 | 35.0 | 0.184211 | 16.0 | 16.0 | 0.026316 | 4.0 | 32.0 | 0.184211 | 16.0 | 16.0 | 0.026316 | 2.0 | 38.0 | 0.789474 | NaN | NaN | NaN | 38 |
| 3 | 5 | 6.0 | 124.0 | 0.044776 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.902985 | 2.0 | 134.0 | 0.940299 | 6.0 | 124.0 | 0.044776 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.955224 | 6.0 | 124.0 | 0.044776 | 2.0 | 134.0 | 0.880597 | 31.0 | 124.0 | 0.029851 | 6.0 | 124.0 | 0.067164 | 3.0 | 134.0 | 0.917910 | 2.0 | 133.0 | 0.917910 | 2.0 | 134.0 | 0.231343 | 6.0 | 124.0 | 0.044776 | 6.0 | 130.0 | 0.186567 | 6.0 | 124.0 | 0.059701 | 2.0 | 133.0 | 0.880597 | 6.0 | 124.0 | 0.044776 | 134 |
| 4 | 6 | 3.0 | 28.0 | 0.039216 | 3.0 | 28.0 | 0.039216 | NaN | NaN | NaN | 2.0 | 50.0 | 0.803922 | 3.0 | 22.0 | 0.058824 | 3.0 | 28.0 | 0.058824 | 2.0 | 49.0 | 0.803922 | 3.0 | 28.0 | 0.039216 | 2.0 | 50.0 | 0.764706 | 3.0 | 28.0 | 0.058824 | 3.0 | 28.0 | 0.058824 | 2.0 | 50.0 | 0.784314 | 2.0 | 49.0 | 0.784314 | 4.0 | 47.0 | 0.215686 | 3.0 | 28.0 | 0.058824 | 3.0 | 28.0 | 0.039216 | 3.0 | 28.0 | 0.039216 | 2.0 | 49.0 | 0.745098 | 3.0 | 28.0 | 0.039216 | 51 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15139 | 21628 | 17.0 | 17.0 | 0.035714 | 17.0 | 17.0 | 0.035714 | 10.0 | 21.0 | 0.428571 | 10.0 | 36.0 | 0.964286 | 10.0 | 17.0 | 0.071429 | 17.0 | 17.0 | 0.035714 | 10.0 | 36.0 | 0.857143 | NaN | NaN | NaN | 10.0 | 36.0 | 0.892857 | 17.0 | 17.0 | 0.035714 | 10.0 | 17.0 | 0.071429 | 10.0 | 36.0 | 0.928571 | 10.0 | 20.0 | 0.392857 | 10.0 | 35.0 | 0.250000 | 17.0 | 17.0 | 0.035714 | 17.0 | 17.0 | 0.035714 | NaN | NaN | NaN | 10.0 | 20.0 | 0.392857 | 17.0 | 17.0 | 0.035714 | 28 |
| 15140 | 21629 | 15.0 | 39.0 | 0.035714 | 15.0 | 39.0 | 0.035714 | 2.0 | 56.0 | 0.857143 | 2.0 | 56.0 | 0.839286 | 15.0 | 39.0 | 0.053571 | 15.0 | 39.0 | 0.053571 | 2.0 | 56.0 | 0.821429 | 15.0 | 15.0 | 0.017857 | 2.0 | 56.0 | 0.821429 | 15.0 | 39.0 | 0.035714 | 3.0 | 39.0 | 0.089286 | 2.0 | 56.0 | 0.839286 | 2.0 | 56.0 | 0.803571 | 2.0 | 56.0 | 0.482143 | 15.0 | 39.0 | 0.035714 | 2.0 | 44.0 | 0.410714 | 3.0 | 39.0 | 0.107143 | 2.0 | 56.0 | 0.767857 | 15.0 | 39.0 | 0.035714 | 56 |
| 15141 | 21630 | 13.0 | 44.0 | 0.069767 | 13.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.930233 | 4.0 | 45.0 | 0.930233 | 5.0 | 44.0 | 0.093023 | 18.0 | 44.0 | 0.046512 | 4.0 | 45.0 | 0.930233 | 5.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.883721 | 18.0 | 44.0 | 0.046512 | 13.0 | 44.0 | 0.069767 | 4.0 | 45.0 | 0.767442 | 4.0 | 45.0 | 0.930233 | 4.0 | 45.0 | 0.906977 | 18.0 | 44.0 | 0.046512 | 4.0 | 44.0 | 0.162791 | 13.0 | 18.0 | 0.046512 | 4.0 | 45.0 | 0.906977 | 13.0 | 44.0 | 0.069767 | 43 |
| 15142 | 21632 | 5.0 | 9.0 | 0.142857 | 5.0 | 5.0 | 0.071429 | 2.0 | 14.0 | 0.857143 | 2.0 | 14.0 | 0.928571 | 5.0 | 13.0 | 0.285714 | 5.0 | 13.0 | 0.285714 | 2.0 | 14.0 | 0.928571 | 5.0 | 9.0 | 0.142857 | 2.0 | 14.0 | 0.857143 | 5.0 | 9.0 | 0.142857 | 5.0 | 9.0 | 0.214286 | 2.0 | 14.0 | 0.928571 | 2.0 | 14.0 | 0.785714 | 4.0 | 14.0 | 0.285714 | 5.0 | 9.0 | 0.142857 | 9.0 | 9.0 | 0.071429 | 5.0 | 9.0 | 0.142857 | 2.0 | 14.0 | 0.785714 | 5.0 | 5.0 | 0.071429 | 14 |
| 15143 | 21633 | 2.0 | 6.0 | 0.157895 | 5.0 | 5.0 | 0.052632 | NaN | NaN | NaN | 2.0 | 19.0 | 0.894737 | 2.0 | 19.0 | 0.368421 | 5.0 | 6.0 | 0.105263 | 2.0 | 18.0 | 0.842105 | 2.0 | 6.0 | 0.157895 | 2.0 | 19.0 | 0.789474 | 5.0 | 5.0 | 0.052632 | 2.0 | 6.0 | 0.157895 | 2.0 | 19.0 | 0.894737 | 2.0 | 19.0 | 0.894737 | 3.0 | 16.0 | 0.263158 | 5.0 | 6.0 | 0.105263 | 5.0 | 5.0 | 0.052632 | 5.0 | 5.0 | 0.052632 | 2.0 | 19.0 | 0.842105 | 5.0 | 5.0 | 0.052632 | 19 |
15144 rows × 59 columns
- We now have 110 -> 59 variables.
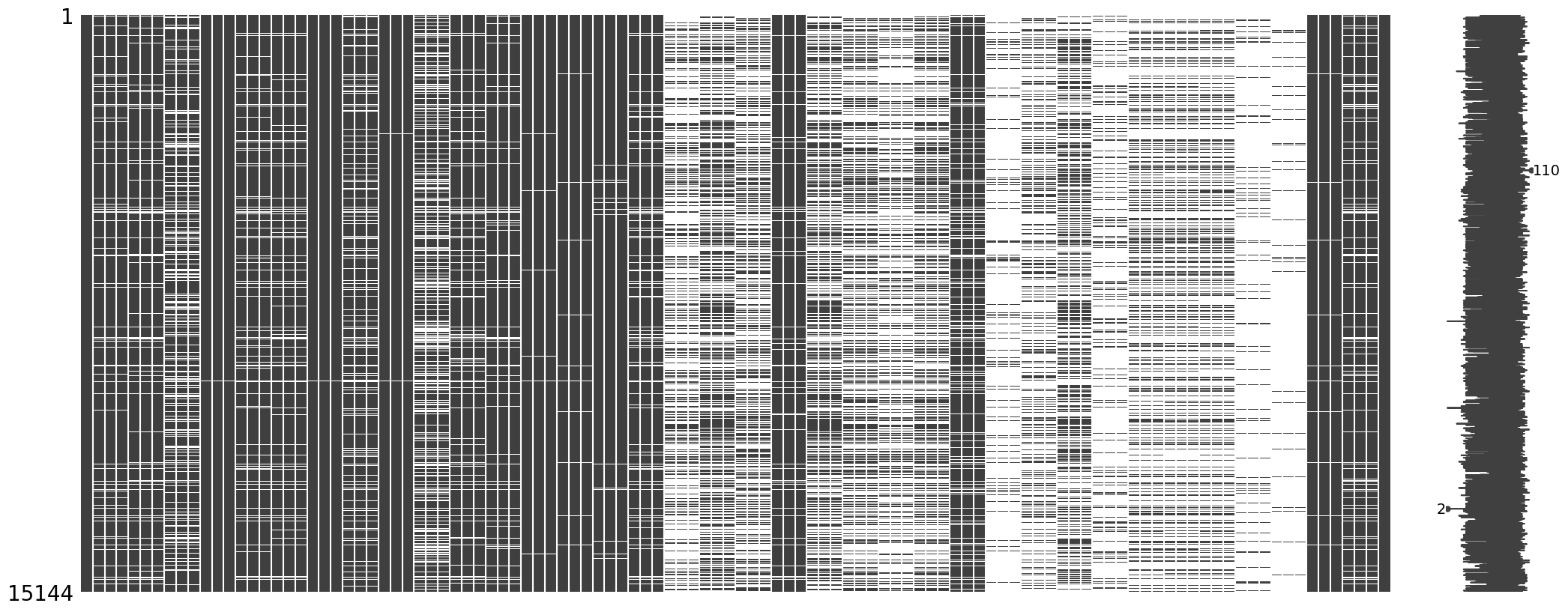
msno.matrix(df_train_measurment_time_info)
<AxesSubplot:>
- There are still some missing values.
- Missing values means that the patient have no record in that variable. So it is reasonable to impute missing value with 0.
df_train_measurment_time_info = df_train_measurment_time_info.fillna(0)
df_test_measurment_time_info = df_test_measurment_time_info.fillna(0)
fig, axes = plt.subplots(1, 2, figsize = (12, 5))
msno.matrix(df_train_measurment_time_info, ax = axes[0])
axes[0].set_title("Train data", fontsize = 15)
msno.matrix(df_test_measurment_time_info, ax = axes[1])
axes[1].set_title("Test data", fontsize = 15)
Text(0.5, 1.0, 'Test data')
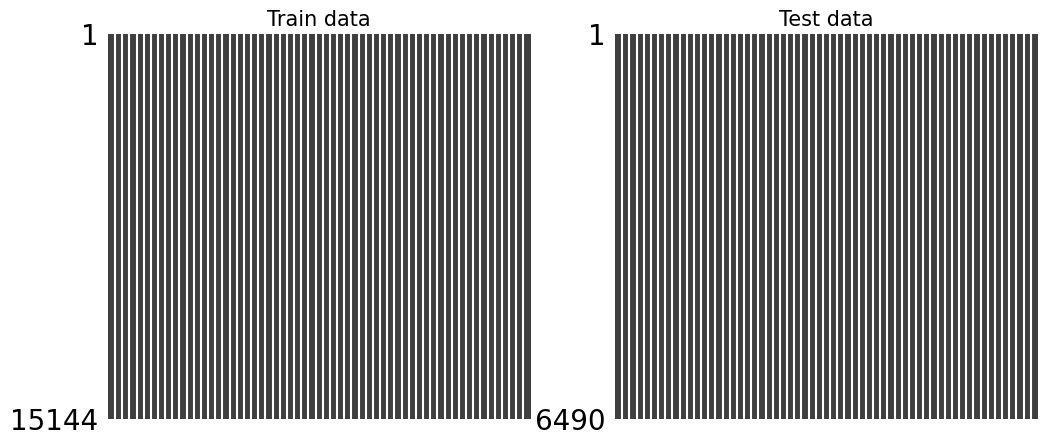
-
Now there is no missing value.
-
Save the tables.
df_train_measurment_time_info.to_csv("../data/df_train_measurment_time_info.csv", index = False)
df_test_measurment_time_info.to_csv("../data/df_test_measurment_time_info.csv", index = False)
3. Summary statistics
msno.matrix(df_train.drop(["Age", "Gender", "Outcome", "ID"], axis = 1))
<AxesSubplot:>

- Because each variable is recorded only when it is measured, there are many missing values.
- Since variables are measured in several time for each patient, this is the dependent dataset. That is, there are correlations between measured values from same patient.
-
So let’s use summary statistics like minimum, maximum, mean, median, standard deviation for each variable instead of several measured variables for each patient.
- Before extract the summary statistics, let’s impute the missing values.
plt.figure(figsize = (15, 5))
sns.boxplot(df_train.drop(["Age", "Gender", "Outcome"], axis = 1).groupby("ID").std(), showfliers = False)
plt.ylabel("Standard Deviation", fontsize = 12)
plt.xlabel("Variables", fontsize = 12)
plt.title("Boxplot of the standard deviation of each patient", fontsize = 15)
plt.xticks(rotation = 90)
plt.show()
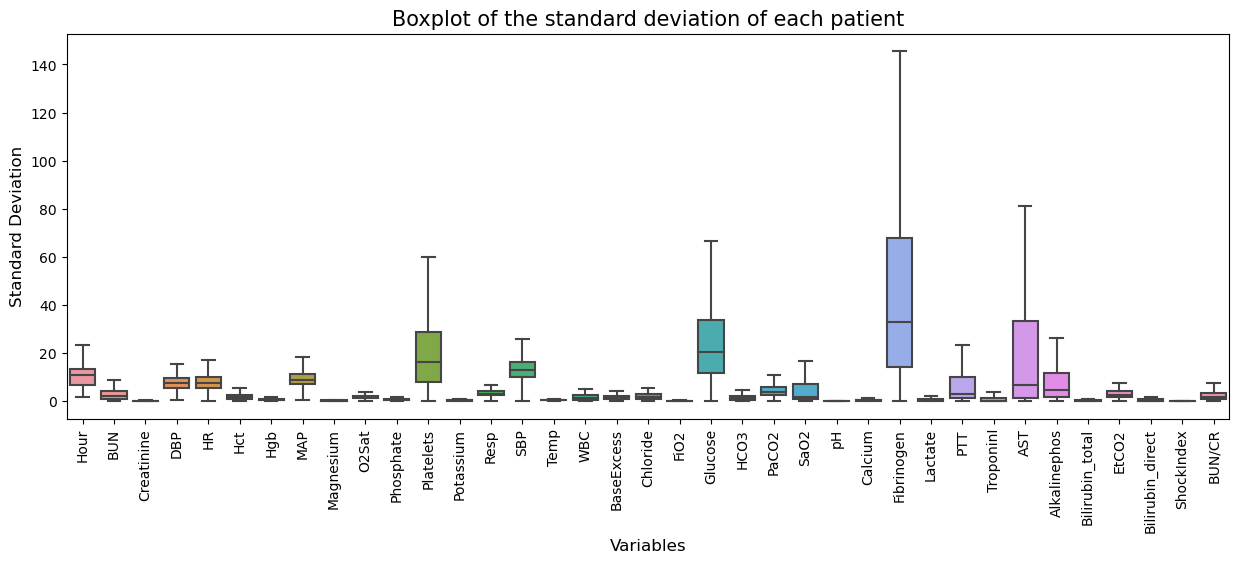
- Above plot shows the standard deviation distribution of each patient’s measurements.
- It can be seen that the standard deviation of patients for almost all variables is very low.
- That is, there is no significant change in the measurement within the same patient.
- So, let’s impute the missing values by the forward filling method which assumed the measurement value remained constant until the next measurement.
target_col_list = ["ID", "Hour", 'BUN', 'Creatinine', 'DBP', 'HR', 'Hct', 'Hgb',
'MAP', 'Magnesium', 'O2Sat', 'Phosphate', 'Platelets',
'Potassium', 'Resp', 'SBP', 'Temp', 'WBC', 'BaseExcess', 'Chloride',
'FiO2', 'Glucose', 'HCO3', 'PaCO2', 'SaO2', 'pH', 'Calcium',
'Fibrinogen', 'Lactate', 'PTT', 'TroponinI', 'AST', 'Alkalinephos',
'Bilirubin_total', 'EtCO2', 'Bilirubin_direct', 'ShockIndex','BUN/CR']
measurement_variable_train = df_train[target_col_list].sort_values(["ID", "Hour"])
measurement_variable_test = df_test[target_col_list].sort_values(["ID", "Hour"])
measurement_variable_train["ID_for_groupby"] = measurement_variable_train.ID
measurement_variable_test["ID_for_groupby"] = measurement_variable_test.ID
measurement_variable_train = measurement_variable_train.groupby("ID_for_groupby").transform(lambda v: v.ffill())
measurement_variable_test = measurement_variable_test.groupby("ID_for_groupby").transform(lambda v: v.ffill())
measurement_variable_train.drop("Hour", axis = 1, inplace = True)
measurement_variable_test.drop("Hour", axis = 1, inplace = True)
msno.matrix(measurement_variable_train)
<AxesSubplot:>
- Now let’s extract the summary statistics of each patient.
# features for train data
grouped_mean = measurement_variable_train.groupby("ID").mean()
grouped_std = measurement_variable_train.groupby("ID").std()
grouped_min = measurement_variable_train.groupby("ID").min()
grouped_max = measurement_variable_train.groupby("ID").max()
for col in grouped_mean.columns:
grouped_mean = grouped_mean.rename(columns = {col: f"{col}_mean"})
grouped_std = grouped_std.rename(columns = {col: f"{col}_std"})
grouped_min = grouped_min.rename(columns = {col: f"{col}_min"})
grouped_max = grouped_max.rename(columns = {col: f"{col}_max"})
df_train_measurement_grouped_stat = measurement_variable_train[["ID"]].drop_duplicates()
df_train_measurement_grouped_stat = df_train_measurement_grouped_stat.merge(grouped_mean, how = "left", on = "ID") \
.merge(grouped_std, how = "left", on = "ID") \
.merge(grouped_min, how = "left", on = "ID") \
.merge(grouped_max, how = "left", on = "ID")
# features for test data
grouped_mean = measurement_variable_test.groupby("ID").mean()
grouped_std = measurement_variable_test.groupby("ID").std()
grouped_min = measurement_variable_test.groupby("ID").min()
grouped_max = measurement_variable_test.groupby("ID").max()
for col in grouped_mean.columns:
grouped_mean = grouped_mean.rename(columns = {col: f"{col}_mean"})
grouped_std = grouped_std.rename(columns = {col: f"{col}_std"})
grouped_min = grouped_min.rename(columns = {col: f"{col}_min"})
grouped_max = grouped_max.rename(columns = {col: f"{col}_max"})
df_test_measurment_grouped_stat = measurement_variable_test[["ID"]].drop_duplicates()
df_test_measurment_grouped_stat = df_test_measurment_grouped_stat.merge(grouped_mean, how = "left", on = "ID") \
.merge(grouped_std, how = "left", on = "ID") \
.merge(grouped_min, how = "left", on = "ID") \
.merge(grouped_max, how = "left", on = "ID")
df_train_measurement_grouped_stat
| ID | BUN_mean | Creatinine_mean | DBP_mean | HR_mean | Hct_mean | Hgb_mean | MAP_mean | Magnesium_mean | O2Sat_mean | Phosphate_mean | Platelets_mean | Potassium_mean | Resp_mean | SBP_mean | Temp_mean | WBC_mean | BaseExcess_mean | Chloride_mean | FiO2_mean | Glucose_mean | HCO3_mean | PaCO2_mean | SaO2_mean | pH_mean | Calcium_mean | Fibrinogen_mean | Lactate_mean | PTT_mean | TroponinI_mean | AST_mean | Alkalinephos_mean | Bilirubin_total_mean | EtCO2_mean | Bilirubin_direct_mean | ShockIndex_mean | BUN/CR_mean | BUN_std | Creatinine_std | DBP_std | HR_std | Hct_std | Hgb_std | MAP_std | Magnesium_std | O2Sat_std | Phosphate_std | Platelets_std | Potassium_std | Resp_std | SBP_std | Temp_std | WBC_std | BaseExcess_std | Chloride_std | FiO2_std | Glucose_std | HCO3_std | PaCO2_std | SaO2_std | pH_std | Calcium_std | Fibrinogen_std | Lactate_std | PTT_std | TroponinI_std | AST_std | Alkalinephos_std | Bilirubin_total_std | EtCO2_std | Bilirubin_direct_std | ShockIndex_std | BUN/CR_std | BUN_min | Creatinine_min | DBP_min | HR_min | Hct_min | Hgb_min | MAP_min | Magnesium_min | O2Sat_min | Phosphate_min | Platelets_min | Potassium_min | Resp_min | SBP_min | Temp_min | WBC_min | BaseExcess_min | Chloride_min | FiO2_min | Glucose_min | HCO3_min | PaCO2_min | SaO2_min | pH_min | Calcium_min | Fibrinogen_min | Lactate_min | PTT_min | TroponinI_min | AST_min | Alkalinephos_min | Bilirubin_total_min | EtCO2_min | Bilirubin_direct_min | ShockIndex_min | BUN/CR_min | BUN_max | Creatinine_max | DBP_max | HR_max | Hct_max | Hgb_max | MAP_max | Magnesium_max | O2Sat_max | Phosphate_max | Platelets_max | Potassium_max | Resp_max | SBP_max | Temp_max | WBC_max | BaseExcess_max | Chloride_max | FiO2_max | Glucose_max | HCO3_max | PaCO2_max | SaO2_max | pH_max | Calcium_max | Fibrinogen_max | Lactate_max | PTT_max | TroponinI_max | AST_max | Alkalinephos_max | Bilirubin_total_max | EtCO2_max | Bilirubin_direct_max | ShockIndex_max | BUN/CR_max | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 12.166250 | 0.700000 | 58.893333 | 89.970476 | 27.213571 | 9.070000 | 75.272381 | NaN | 97.984762 | NaN | NaN | 4.471250 | 15.700476 | 115.151905 | 36.694000 | 8.036250 | 0.990000 | 107.468750 | 0.400000 | 125.801429 | 25.825000 | 37.500000 | 99.281429 | 7.443333 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.789651 | 16.942857 | 0.123744 | 0.000000 | 8.480952 | 4.234100 | 1.497745 | 0.056569 | 7.690843 | NaN | 1.062688 | NaN | NaN | 0.003536 | 1.964122 | 10.043408 | 0.491993 | 0.045962 | 0.000000 | 0.003536 | 0.000000 | 15.032899 | 0.014142 | 0.000000 | 0.635014 | 0.030551 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.088723 | 0.000000 | 11.86 | 0.70 | 46.93 | 74.78 | 25.96 | 9.05 | 60.12 | NaN | 95.93 | NaN | NaN | 4.47 | 11.41 | 99.02 | 35.61 | 8.02 | 0.99 | 107.46 | 0.40 | 107.10 | 25.79 | 37.50 | 96.51 | 7.31 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.614563 | 16.942857 | 12.21 | 0.70 | 79.91 | 95.72 | 28.89 | 9.21 | 90.60 | NaN | 100.06 | NaN | NaN | 4.48 | 19.98 | 132.76 | 37.41 | 8.15 | 0.99 | 107.47 | 0.40 | 137.31 | 25.83 | 37.50 | 99.42 | 7.45 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.966673 | 16.942857 |
| 1 | 3 | 9.179091 | 0.563636 | 67.792982 | 82.789474 | 30.123636 | 10.530909 | 76.083684 | 2.531176 | 96.276842 | 3.800000 | 212.173636 | 4.121765 | 17.965439 | 120.228070 | 36.999636 | 11.948864 | 2.123000 | 109.641136 | 0.412973 | 126.724314 | 25.850455 | 44.505600 | 78.998108 | 7.408200 | 8.430196 | 175.62 | 1.464400 | 37.180909 | NaN | NaN | NaN | NaN | NaN | NaN | 0.701669 | 16.400303 | 2.111585 | 0.048661 | 21.534443 | 14.053193 | 3.011037 | 1.034972 | 10.881973 | 0.383639 | 1.951691 | 0.000000 | 29.228191 | 0.386175 | 5.022348 | 15.108003 | 0.789538 | 2.762756 | 2.146345 | 6.127336 | 0.084422 | 32.436454 | 1.442959 | 4.729308 | 15.239642 | 0.033178 | 0.122466 | 36.734541 | 0.664272 | 12.378049 | NaN | NaN | NaN | NaN | NaN | NaN | 0.165108 | 3.771555 | 7.72 | 0.50 | 34.82 | 60.35 | 24.46 | 8.05 | 52.87 | 2.00 | 91.13 | 3.80 | 177.48 | 3.61 | 9.64 | 91.85 | 35.80 | 6.82 | -2.86 | 104.39 | 0.39 | 50.39 | 25.23 | 31.13 | 65.03 | 7.37 | 8.21 | 161.19 | 0.88 | 23.81 | NaN | NaN | NaN | NaN | NaN | NaN | 0.494388 | 12.866667 | 13.76 | 0.60 | 97.18 | 111.08 | 40.50 | 13.51 | 109.62 | 3.11 | 100.17 | 3.80 | 281.18 | 5.04 | 34.05 | 160.07 | 38.28 | 13.94 | 12.85 | 117.54 | 0.75 | 169.06 | 29.13 | 49.32 | 98.44 | 7.51 | 8.51 | 267.01 | 3.63 | 52.77 | NaN | NaN | NaN | NaN | NaN | NaN | 1.118672 | 22.933333 |
| 2 | 4 | NaN | 0.750000 | 77.953514 | 106.853243 | 42.500000 | 14.200000 | 104.294324 | NaN | 95.424054 | NaN | NaN | 4.200000 | 23.717297 | 146.205676 | 36.891944 | 15.000000 | NaN | NaN | NaN | 212.999143 | NaN | NaN | NaN | NaN | 9.300000 | NaN | NaN | NaN | 0.07 | NaN | NaN | NaN | NaN | NaN | 0.726613 | NaN | NaN | 0.000000 | 9.103789 | 8.915002 | 0.000000 | 0.000000 | 7.853111 | NaN | 1.396676 | NaN | NaN | 0.000000 | 6.562330 | 12.773692 | 0.320114 | 0.000000 | NaN | NaN | NaN | 63.719123 | NaN | NaN | NaN | NaN | 0.000000 | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.075166 | NaN | NaN | 0.75 | 62.68 | 85.62 | 42.50 | 14.20 | 88.96 | NaN | 92.91 | NaN | NaN | 4.20 | 15.51 | 122.67 | 36.32 | 15.00 | NaN | NaN | NaN | 124.72 | NaN | NaN | NaN | NaN | 9.30 | NaN | NaN | NaN | 0.07 | NaN | NaN | NaN | NaN | NaN | 0.619538 | NaN | NaN | 0.75 | 99.83 | 129.44 | 42.50 | 14.20 | 126.46 | NaN | 98.13 | NaN | NaN | 4.20 | 35.39 | 173.46 | 37.30 | 15.00 | NaN | NaN | NaN | 304.73 | NaN | NaN | NaN | NaN | 9.30 | NaN | NaN | NaN | 0.07 | NaN | NaN | NaN | NaN | NaN | 0.905600 | NaN |
| 3 | 5 | 12.160155 | 0.712868 | 75.141654 | 75.061504 | 42.821163 | 14.231240 | 99.213158 | 1.849922 | 96.837895 | 2.534574 | 83.771154 | 3.808992 | 22.834015 | 143.588947 | 37.342256 | 10.195039 | NaN | NaN | 0.665977 | 121.621318 | NaN | 38.225208 | 97.278942 | 7.341635 | 6.567364 | NaN | 0.855079 | 29.580000 | 0.01 | 36.000 | 58.00 | 1.3 | 30.981461 | NaN | 0.538676 | 17.183755 | 3.256115 | 0.049768 | 17.953965 | 6.938271 | 3.263822 | 1.568429 | 23.095705 | 0.251668 | 1.794488 | 0.544217 | 16.726549 | 0.379603 | 5.684222 | 26.486614 | 0.554015 | 1.514849 | NaN | NaN | 0.244319 | 11.195427 | NaN | 6.224882 | 2.014454 | 0.073816 | 2.751945 | NaN | 0.079268 | 0.000000 | 0.0 | 0.000000 | 0.0 | 0.0 | 4.982443 | NaN | 0.092515 | 4.809614 | 7.85 | 0.62 | 46.62 | 60.29 | 36.22 | 11.77 | 61.15 | 1.49 | 90.87 | 2.08 | 73.72 | 3.21 | 9.53 | 89.44 | 35.94 | 7.41 | NaN | NaN | 0.39 | 96.87 | NaN | 24.69 | 92.83 | 7.25 | 0.93 | NaN | 0.66 | 29.58 | 0.01 | 36.00 | 58.00 | 1.3 | 25.1 | NaN | 0.359089 | 10.101266 | 15.25 | 0.79 | 119.31 | 90.65 | 47.05 | 16.28 | 159.52 | 2.28 | 100.11 | 3.66 | 130.94 | 4.42 | 30.98 | 207.72 | 38.26 | 12.45 | NaN | NaN | 0.99 | 142.74 | NaN | 51.11 | 99.24 | 7.51 | 8.73 | NaN | 0.89 | 29.58 | 0.01 | 36.00 | 58.00 | 1.3 | 45.94 | NaN | 0.831299 | 22.177419 |
| 4 | 6 | 26.554082 | 0.467959 | NaN | 61.949000 | 29.122245 | 9.604286 | 82.788000 | 2.188980 | 97.344800 | 3.171837 | 395.151020 | 4.005306 | 18.338800 | 134.208600 | 37.135417 | 14.732653 | NaN | 113.110408 | NaN | 80.855102 | 24.154694 | NaN | NaN | NaN | 8.218980 | 611.00 | NaN | 38.032041 | NaN | 24.326 | 70.18 | 0.3 | NaN | NaN | 0.471018 | 57.793576 | 6.884190 | 0.146472 | NaN | 5.563790 | 0.869107 | 0.237171 | 16.465945 | 0.196980 | 2.092771 | 0.585888 | 20.687366 | 0.308289 | 2.266595 | 20.055612 | 0.590178 | 2.146741 | NaN | 3.444620 | NaN | 6.192235 | 1.964747 | NaN | NaN | NaN | 0.196980 | 0.000000 | NaN | 5.530593 | NaN | 0.846223 | 0.0 | 0.0 | NaN | NaN | 0.086874 | 3.421685 | 19.60 | 0.32 | NaN | 50.53 | 28.47 | 9.39 | 51.80 | 1.99 | 92.95 | 2.58 | 377.25 | 3.63 | 13.90 | 92.72 | 35.95 | 12.60 | NaN | 109.77 | NaN | 74.60 | 22.17 | NaN | NaN | NaN | 8.02 | 611.00 | NaN | 33.73 | NaN | 23.91 | 70.18 | 0.3 | NaN | NaN | 0.322052 | 54.475410 | 33.23 | 0.61 | NaN | 73.38 | 30.55 | 9.99 | 130.20 | 2.38 | 100.17 | 3.74 | 430.45 | 4.44 | 22.63 | 172.93 | 38.19 | 17.44 | NaN | 116.59 | NaN | 86.86 | 26.06 | NaN | NaN | NaN | 8.41 | 611.00 | NaN | 48.15 | NaN | 25.99 | 70.18 | 0.3 | NaN | NaN | 0.758520 | 61.250000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15139 | 21628 | 14.000000 | 0.700000 | 49.540741 | 103.457778 | 32.625185 | 11.900000 | 72.065926 | NaN | 94.868148 | NaN | 178.000000 | 3.947778 | 22.161481 | 103.968148 | 36.998889 | 16.200000 | NaN | 104.014074 | NaN | 130.000000 | 20.000000 | NaN | NaN | NaN | NaN | NaN | NaN | 29.100000 | NaN | NaN | NaN | NaN | NaN | NaN | 1.038000 | 20.000000 | 0.000000 | 0.000000 | 4.899731 | 4.839157 | 0.593946 | 0.000000 | 10.556669 | NaN | 1.951857 | NaN | 0.000000 | 0.013397 | 5.525944 | 7.127929 | 0.508386 | 0.000000 | NaN | 1.884551 | NaN | 0.000000 | 0.000000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.000000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.071324 | 0.000000 | 14.00 | 0.70 | 43.00 | 94.72 | 31.64 | 11.90 | 40.90 | NaN | 88.11 | NaN | 178.00 | 3.94 | 10.79 | 92.94 | 36.21 | 16.20 | NaN | 102.92 | NaN | 130.00 | 20.00 | NaN | NaN | NaN | NaN | NaN | NaN | 29.10 | NaN | NaN | NaN | NaN | NaN | NaN | 0.846464 | 20.000000 | 14.00 | 0.70 | 64.98 | 112.09 | 32.97 | 11.90 | 86.11 | NaN | 97.84 | NaN | 178.00 | 3.97 | 32.90 | 126.55 | 37.67 | 16.20 | NaN | 107.14 | NaN | 130.00 | 20.00 | NaN | NaN | NaN | NaN | NaN | NaN | 29.10 | NaN | NaN | NaN | NaN | NaN | NaN | 1.081773 | 20.000000 |
| 15140 | 21629 | 23.535714 | 1.080000 | 54.228182 | 90.621455 | 27.461905 | 9.251905 | 83.219091 | 2.510000 | 99.656364 | NaN | 61.812857 | 4.172222 | 21.536545 | 135.544545 | 36.688909 | 8.975714 | NaN | NaN | 0.400000 | 118.397818 | NaN | 38.955185 | 98.352222 | 7.324815 | 7.520370 | NaN | 2.288519 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.678145 | 21.818146 | 1.993464 | 0.105183 | 4.459638 | 2.390373 | 0.436992 | 0.198718 | 8.077108 | 0.000000 | 0.625843 | NaN | 2.579482 | 0.081000 | 3.855742 | 17.470132 | 0.372527 | 0.766332 | NaN | NaN | 0.000000 | 20.257351 | NaN | 1.630796 | 0.209516 | 0.014108 | 3.382747 | NaN | 0.042445 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.080650 | 0.271558 | 21.83 | 0.99 | 44.44 | 82.96 | 26.98 | 9.09 | 66.31 | 2.51 | 97.02 | NaN | 58.87 | 4.07 | 9.88 | 100.25 | 35.29 | 8.32 | NaN | NaN | 0.40 | 80.11 | NaN | 31.73 | 98.28 | 7.32 | 0.95 | NaN | 2.14 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.535106 | 21.508333 | 25.81 | 1.20 | 63.44 | 97.10 | 27.98 | 9.49 | 97.45 | 2.51 | 100.07 | NaN | 64.02 | 4.33 | 28.28 | 169.63 | 37.03 | 9.85 | NaN | NaN | 0.40 | 162.05 | NaN | 39.45 | 99.07 | 7.37 | 9.78 | NaN | 2.31 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.838504 | 22.050505 |
| 15141 | 21630 | 42.466667 | 4.120606 | 62.765714 | 94.085476 | 30.212195 | 10.845714 | 83.868810 | 2.177317 | 97.519524 | 4.575152 | 296.011429 | 4.180000 | 11.622857 | 123.615238 | 36.859048 | 21.472857 | -4.222381 | 104.863571 | NaN | 111.255238 | 18.461212 | 36.479762 | 53.000000 | 7.349762 | 7.987879 | NaN | 1.903636 | 27.370000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.770192 | 10.293502 | 2.893742 | 0.178745 | 4.091623 | 3.689259 | 2.089079 | 0.456342 | 7.211074 | 0.122373 | 1.486316 | 0.093378 | 28.927859 | 0.144005 | 2.910789 | 12.316088 | 0.517663 | 1.458195 | 1.489483 | 0.794664 | NaN | 27.694204 | 0.834175 | 1.216553 | NaN | 0.022684 | 0.029129 | NaN | 0.207542 | 0.000000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.079790 | 0.297607 | 31.82 | 3.47 | 48.85 | 79.82 | 26.22 | 9.23 | 63.21 | 2.11 | 92.95 | 4.40 | 193.59 | 4.11 | 2.79 | 96.03 | 35.53 | 16.31 | -5.03 | 102.05 | NaN | 89.49 | 17.06 | 34.89 | 53.00 | 7.32 | 7.92 | NaN | 1.42 | 27.37 | NaN | NaN | NaN | NaN | NaN | NaN | 0.601054 | 9.170029 | 43.56 | 4.19 | 70.04 | 97.33 | 31.75 | 10.97 | 97.55 | 2.41 | 100.02 | 4.82 | 303.89 | 4.50 | 18.11 | 150.01 | 37.68 | 21.87 | 0.04 | 105.08 | NaN | 170.49 | 20.94 | 39.11 | 53.00 | 7.40 | 8.00 | NaN | 1.99 | 27.37 | NaN | NaN | NaN | NaN | NaN | NaN | 0.987712 | 10.396181 |
| 15142 | 21632 | 80.246000 | 1.800000 | 65.857692 | 96.119231 | 33.046000 | 10.938000 | 77.470769 | 1.994000 | 98.196154 | 4.474000 | 290.938000 | 6.108000 | 30.662308 | 92.554615 | 38.930000 | 43.398000 | -5.921000 | 108.630000 | 0.990000 | 239.020000 | 16.376000 | 37.802000 | 88.181000 | 7.305000 | 7.544000 | NaN | 2.010000 | 76.500000 | NaN | 519.000 | 56.00 | 0.3 | NaN | NaN | 1.042062 | 43.911111 | 1.037960 | 0.000000 | 5.216346 | 4.880952 | 1.442338 | 0.413247 | 5.570530 | 0.005164 | 2.724957 | 0.495742 | 9.796066 | 0.242981 | 4.044771 | 5.741656 | 0.615305 | 2.153379 | 1.176997 | 0.490578 | 0.031623 | 0.000000 | 0.562874 | 5.151450 | 13.689373 | 0.041966 | 0.056804 | NaN | 0.144914 | 0.000000 | NaN | 0.000000 | 0.0 | 0.0 | NaN | NaN | 0.081292 | 0.000000 | 79.04 | 1.80 | 58.65 | 89.91 | 31.10 | 10.37 | 66.00 | 1.99 | 92.90 | 4.09 | 283.35 | 5.92 | 25.36 | 85.35 | 38.16 | 41.73 | -7.12 | 108.06 | 0.90 | 239.02 | 15.94 | 34.10 | 68.35 | 7.25 | 7.50 | NaN | 1.80 | 76.50 | NaN | 519.00 | 56.00 | 0.3 | NaN | NaN | 0.910573 | 43.911111 | 81.05 | 1.80 | 75.78 | 102.35 | 34.38 | 11.31 | 86.98 | 2.00 | 100.25 | 5.05 | 302.32 | 6.42 | 38.91 | 101.49 | 39.68 | 45.90 | -3.91 | 109.01 | 1.00 | 239.02 | 17.03 | 47.03 | 97.26 | 7.36 | 7.61 | NaN | 2.10 | 76.50 | NaN | 519.00 | 56.00 | 0.3 | NaN | NaN | 1.194004 | 43.911111 |
| 15143 | 21633 | 93.810556 | 1.200000 | NaN | 90.822778 | 29.042222 | 10.201333 | 72.558333 | 1.685000 | 98.795556 | 3.200000 | 167.000000 | 4.187222 | 17.545000 | 108.194444 | 36.319412 | 16.992000 | NaN | 111.816111 | NaN | 90.000000 | 16.867778 | NaN | NaN | NaN | 8.100000 | NaN | NaN | 30.400000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.868208 | 77.541667 | 1.835882 | 0.000000 | NaN | 6.026391 | 2.952490 | 0.033566 | 20.329933 | 0.009852 | 3.708108 | 0.000000 | 0.000000 | 0.594928 | 2.099718 | 24.719105 | 0.204648 | 0.007746 | NaN | 0.403195 | NaN | 0.000000 | 0.413245 | NaN | NaN | NaN | 0.000000 | NaN | NaN | 0.000000 | NaN | NaN | NaN | NaN | NaN | NaN | 0.135540 | 0.000000 | 93.01 | 1.20 | NaN | 76.23 | 23.18 | 10.08 | 39.45 | 1.68 | 84.14 | 3.20 | 167.00 | 3.91 | 14.91 | 85.04 | 36.01 | 16.99 | NaN | 110.94 | NaN | 90.00 | 15.97 | NaN | NaN | NaN | 8.10 | NaN | NaN | 30.40 | NaN | NaN | NaN | NaN | NaN | NaN | 0.583887 | 77.541667 | 97.80 | 1.20 | NaN | 99.29 | 31.55 | 10.21 | 147.23 | 1.71 | 100.32 | 3.20 | 167.00 | 5.48 | 22.79 | 170.05 | 36.55 | 17.02 | NaN | 112.01 | NaN | 90.00 | 17.05 | NaN | NaN | NaN | 8.10 | NaN | NaN | 30.40 | NaN | NaN | NaN | NaN | NaN | NaN | 1.104143 | 77.541667 |
15144 rows × 145 columns
msno.matrix(df_train_measurement_grouped_stat)
<AxesSubplot:>

- There are still some missing values.
- Drop the variables with more than 20% missing values.
target_col = df_train_measurement_grouped_stat.isna().sum()[(df_train_measurement_grouped_stat.isna().sum() / df_train_measurement_grouped_stat.shape[0]) < 0.2].reset_index()["index"]
df_train_measurement_grouped_stat = df_train_measurement_grouped_stat[target_col]
df_test_measurment_grouped_stat = df_test_measurment_grouped_stat[target_col]
df_train_measurement_grouped_stat
-
Now we have 145 -> 77 columns.
- There are some relationships between vital signs and laboratory variables, i.e.
- DBP < MAP < SBP.
- $ \widehat{MAP} = DBP + 1/3(SBP - DBP)$
- That is, missing values can be infered by combination of other variables.
- So, let’s impute the missing values by iterative impute method.
imp = IterativeImputer(max_iter = 10, random_state=42)
imp.fit(df_train_measurement_grouped_stat)
IterativeImputer(random_state=42)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
IterativeImputer(random_state=42)
col_names = df_train_measurement_grouped_stat.columns
df_train_measurement_grouped_stat = pd.DataFrame(imp.transform(df_train_measurement_grouped_stat),
columns = col_names)
df_test_measurment_grouped_stat = pd.DataFrame(imp.transform(df_test_measurment_grouped_stat),
columns = col_names)
fig, axes = plt.subplots(1, 2, figsize = (12, 5))
msno.matrix(df_train_measurement_grouped_stat, ax = axes[0])
axes[0].set_title("Train data", fontsize = 15)
msno.matrix(df_test_measurment_grouped_stat, ax = axes[1])
axes[1].set_title("Test data", fontsize = 15)
Text(0.5, 1.0, 'Test data')
-
Now there is no missing value.
-
Save the tables.
df_train_measurement_grouped_stat.to_csv("../data/df_train_measurement_grouped_stat.csv", index = False)
df_test_measurment_grouped_stat.to_csv("../data/df_test_measurment_grouped_stat.csv", index = False)
4. Summary
- Make 2 features based on the research (Henry, K. E., Hager, D. N., Pronovost, P. J., and Saria, S. A targeted real-time early warning score (trewscore) for septic shock. Science Translational Medicine 7, 299 (2015), 299ra122–299ra122.)
- ShockIndex(t) = $\frac{\text{HR(t)}}{SBP(t)}$
- BUN/CR(t) = $\frac{\text{BUN(t)}}{Creatinine(t)}$
- Incorporate the measurment time information of each variables:
- Beginning time: Time of the first measurment
- End time: Time of the last measurment
- Frequency: Ratio between the total number of the measurment and the maximum hour of the patient
-
Also merge the information that shows how long each patient has stayed (the maximum hour of each patient).
- Because each variable is recorded only when it is measured, there are many missing values.
- Since variables are measured in several time for each patient, this is the dependent dataset. That is, there are correlations between measured values from same patient.
- So use summary statistics like minimum, maximum, mean, and standard deviation for each variable instead of several measured variables for each patient.